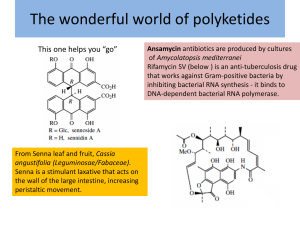
DNA Lecture
advertisement

Microbial Community Analysis With thanks to: Boris Wawrik, Ph.D. Jerome Kukor, Ph.D. Lee Kerkhof, Ph.D. Microbial ecology Long term goals: To gain a better understanding of the ecology of important microorganisms in environmental samples Questions we ask: Which bacteria are present in a sample? How many different types? Which bacteria are active and growing? What’s the ecology of microorganisms in the context of their environment ? How can we apply this knowledge (e.g. bioremediation, fermentation)? Traditional Approach Culture Organisms Isolate Pure Cultures Study Metabolism of Cultures Direct plating of seawater One ml of seawater typically yields 102-103 colonies This is a low number => bacteria were not considered important in the marine environment Electron microscopy Electron microscopy suggested much higher bacterial abundance in the marine environment (1920s) Here are some reasons why 1) Most cells are dead. 2) Many bacteria can not grow on the media. 3) In principle the media components are fine, the concentration is off. 4) The cells grow too slowly for you to assay. 5) Many cells become inactivated by fast growing colonies producing inhibitors. 6) Changes in conditions inhibit growth (e.g. temperature, pressure, placement on agar) 7) Cells clump. 8) Cells stick to pipettes, dilution tubes, sampling gear. The Revolution : Polymerase Chain Reaction (PCR) C G C C T T G 1 40 A A G G C C G C C G 1 50 T C T C G T G C C G 1 60 G T C G T T C T G G C G 1 70 G G T G C C C G 1 80 A C C C G T G C G C G T T G 1 90 A T G T A G T C G 2 00 A T G T C C T C G 2 10 G G G G C What can molecules tell us ? The Central Dogma of Molecular Biology DNA RNA Who is there ? Who is not there ? What functional genes are there ? BUT can not tell you who is active Who is active ? Who is expressing genes? Protein Enzyme activity Rate measurement (e.g. primary production by 14C carbon fixation) Traditional vs. culture independent methods Dunbar et al., AEM No. 4, Vol 65, 1999, pp. 1662-69 known novel Microbial Life, BOX 17.5 Why use Ribosomal RNA Genes? 1. Everybody’s got ‘em 2. All perform the same function--protein synthesis 3. High homology--good for probing or PCR. 4. Good for telling us big picture lineages. 5. Many new rapid molecular biological methods to detect (Pauling and Zuckerkandl-1965; Woese, 1987) Why is the small subunit rRNA gene so useful ? Conserved in parts – highly variable in other parts. Thus it a very good phylogenetic marker VERY large database of sequences Cell have many ribosomes which can be targeted with probes (e.g. FISH, &TRFLP) for community analysis 16S rRNA gene sequencing is now the gold standard for community analysis Primer design: degenerate PCR Conserved sequence shared by all species * * * * * * Ambiguities in the sequence 5’-TWCGTSGARCTGCACGGVACCGGYAC-3’ IUPAC degeneracies: W = A or T V = C or G or A S = G or C Y = C or T R = A or G 2*2*2*3*2 = 48 different primers sequences Some caveats: Not all methods yield the same results Different samples require different extraction methods It is best to try several methods and determine effort, yield, and purity Most people nowadays opt for extractions kits, because they are simple, rapid, reproducible and reasonable cheap Biggest problems: PCR inhibitors that co-extract Low DNA yields (e.g. clay) Who are all these uncultivated bacteria ? There are regions that are highly similar among all bacteria These regions can be used to design universal 16S PCR primers Using these primers we can amplify the 16S sequences from a natural population This mixture of PCR products can be cloned and the inserts from individual colonies sequenced (Woese, Giovannoni, Ward, Stahl, Pace and others – late 1980s and early 1990s) Microbial Life-’02 Perry et al. How do we estimate bacterial community composition ? 99 Streptomyces nodosus AAK73514 I 99 35 Streptomyces nodosus AAK73514 V Streptomyces noursei AF263912 IV 99 Streptomyces noursei AF263912 Streptomyces natalensis AJ278573 V Streptomyces natalensis AJ278573 III Streptomyces natalensis AJ278573 IV Streptomyces natalensis AJ278573 I Streptomyces noursei AF263912 I Streptomyces natalensis AJ278573 VI 4 Streptomyces sp. FR-008 AY310323 I 6 Streptomyces sp. FR-008 AY310323 VI 32 Streptomyces sp. FR-008 AY310323 V Streptomyces natalensis AJ278573 II 99 Streptomyces nodosus AAK73514 III 43 66 Streptomyces nodosus AAK73514 VI 92 Streptomyces nodosus AAK73514 II Streptomyces nodosus AAK73514 IV 99 1 Streptomyces noursei AF263912 III 56 Streptomyces noursei AF263912 II 64 Streptomyces noursei AF263912 V 99 Streptomyces noursei AF263912 VI 99 Streptomyces nanchangensis AF521085 II 54 Streptomyces cinnamonensis AF440781 II 11 Streptomyces natalensis AJ132222 I Streptomyces natalensis AJ132222 II 25 90 Streptomyces caelestis AF016585 II 99 Streptomyces caelestis AF016585 III Streptomyces hygroscopicus AAF86396 V 15 Streptomyces hygroscopicus AAF86396 IV 53 Streptomyces hygroscopicus AAF86396 II 87 Streptomyces hygroscopicus AAF86396 III 99 Streptomyces venezuelae T17409 II 20 Streptomyces avermitilis BAB69303 II Streptomyces sp. HK803 AAQ84157 II 90 Streptomyces sp. HK803 AAQ84157 I 19 Streptomyces sp. FR-008 AY310323 II 87 98 Streptomyces sp. FR-008 AY310323 III 73 Streptomyces sp. FR-008 AY310323 IV 44 Streptomyces natalensis AJ132222 III Streptomyces natalensis AJ132222 IV 99 Streptomyces avermitilis BAB69303 III 97 Streptomyces halstedii BAD08359 III 57 Streptomyces halstedii BAD08359 II 46 Streptomyces noursei AF263912 VII 85 Streptomyces fradiae AAB66504 II 77 Streptomyces antibioticus AF220951 II 94 Streptomyces antibioticus AF220951 III 99 Streptomyces venezuelae T17409 III Streptomyces venezuelae T17409 I Streptomyces coelicolor A3 NP 733695 I Streptomyces antibioticus AF220951 I 39 99 35 Streptomyces nanchangensis AF521085 I 23 Streptomyces cinnamonensis AF440781 I 35 Streptomyces griseoruber AY196994 I 99 Micromonospora griseorubida AB017641 I 53 Micromonospora griseorubida AB089954 I 16 51 Streptomyces fradiae AAB66504 I Streptomyces caelestis AF016585 I Saccharopolyspora erythraea AY330485 I 9 Saccharopolyspora spinosa AY466441 I 7 Streptomyces avermitilis AB070949 I 11 Streptomyces avermitilis AB070949 99 Streptomyces avermitilis BAB69303 I 22 56 99 2 96 C G C C T T G A A G G C C G C C G T C T C G T G C C G G T C G T T C T G G C G G G T G C C C G AC C C G T G C G C G T T G AT G T A G T C G A T G T C C T C G G G G G C 140 150 160 170 180 190 200 210 DNA extraction Primer design and PCR Sequencing Phylogenetic analysis 0.05 Library screening TA cloning Comparison to other samples – hypothesis testing Examples of what you can do with 16S PCR technology DGGE TRFLP SIP FISH DGGE (Denaturing Gradient Gel Electrophoresis) simple complex PCR products of mixed communities are loaded on a gel with a gradient of denaturant Typically 20-80% formamide double stranded DNA will run down the gel until it melts Melting determined by sequence and GC content Different sequences migrate different distances 20% 80% You obtain a ‘barcode’ of the community DGGE (cont.) Advantages Can cut individual bands and clone or sequence them Can detect very small differences in DNA sequences Disadvantages High complexity samples give smears Requires specialized gel rig Acryl-amide is highly toxic TRFLP (Terminal Restriction Fragment Length Polymorphism) cut with 4bp RE FU fragment size Mixed population is amplified using a 16S primer with a fluorescent tag PCR product is cut with a 4bp cutting restriction endonuclease Different sequences will give different length fragments Sample is injected into a capillary sequencer to sort fragments by size TRFLP (cont.) Advantages Very sensitive Fast, easy and cheap Disadvantages Can NOT cut bands to get sequence data Requires capillary sequencer Hard to distinguish noise from little peaks sometimes PCR is inherently NOT quantitative Amplification of some sequences maybe be sub-optimal Reaction kinetics Primer binding Secondary structure of template Amplification tends to lead to a 1:1 product ratio regardless of the starting DNA ratios Amplification of low abundance templates in a mixed template experiment will often be suppressed PCR can produce erroneous sequences Mis-incorporation of nucleotides by TAQ polymerase Formation of chimeric sequences LIBRARY CONTENT CANOT BE USED TO CALCULTE DIVERSITY INDICES Many questions in ecology involve determining the active portion of a community Many species may be present but only a few might be active If you are looking for a functional gene, only some of the bacteria that contain this gene may be involved in actual substrate transformation Among the active ones, who is most dominant/active? Which bacteria are stimulated by a treatment (treatments may not kill other bacteria and 16S can detect them, although they are no longer active)? Stable isotope probing Bacterial population 13C apple pie A population is grown on a substrate that contains 13C carbon Cells that eat the 13C labeled substrate will incorporate it into their DNA. Dormant cells will not DNA extracted and heavy (13C containing) DNA is separated from light (only 12C containing) DNA by CsCl density gradient centrifugation The heavy band is isolated and the community analyzed by PCR – TA cloning approach + grow on labeled substrate DNA 13C DNA centrifugation CsCl gradient extract DNA/RNA 12C WS01ST2CH16 69 WS01ST3CH19 WS01ST3CH4 WS01ST2CH36 WS01ST5CH4 WS01ST7CH32 WS01ST7CH38 WS01ST6CH15 WS01ST6CH37 52 WS01ST6CH17 62 WS01ST6CH2 WS01ST7CH7 WS01ST6CH19 WS01ST7CH4 WS01ST5CH1 WS01ST6CH6 WS01ST2SY14 WS01ST2SY18 77 WS01ST6CH21 WS01ST5CH11 WS01ST6CH22 Diatoms Cylindrotheca sp WS01ST4CH12 WS01ST7CH1 WS01ST5CH14 WS01ST2SY17 Detonula confervacea Odontella sinensis WS01ST4CH15 WS01ST2CH2 WS01ST4CH20 95 WS01ST7CH27 WS01ST4CH36 55 WS01ST7CH18 WS01ST6CH25 WS01ST2CH4 WS01ST4CH31 WS01ST7CH19 Skeletonema costatum WS01ST4CH14 WS01ST6CH1 54 Phaeodactylum tricornutum 50 Bollidomonas pacifica WS01ST7CH3 Bollidophytes Bollidomonas mediterranea WS01ST4CH16 Dictyochophyceae Pseudopedinella elastica 76 100 WS01ST4CH4 67 WS01ST6CH33 Xanthophyceae Heterococcus caespitosus 74 WS01ST1CH14 99 WS01ST3CH27 74 WS01ST8CH5 Eustigmatophytes Nannochloropsis CCMP533 WS01ST1CH4 100 WS01ST1CH9 WS01ST3CH8 WS01ST1CH1 98 WS01ST3CH3 WS01ST1CH8 WS01ST3CH36 WS01ST8CH16 WS01ST1CH33 WS01ST8CH3 97 WS01ST8CH4 P994AH1 WS01ST8CH2 52 97 WS01ST8CH9 P994BH5 WS01ST1CH3 WS01ST6CH16 WS01ST5CH18 WS01ST1CH5 WS01ST2CH10 Prymnesiophytes Chrysochromulina hirta WS01ST1CH27 96 WS01ST3CH24 WS01ST4CH34 97 WS01ST7CH21 99 WS01ST8CH14 WS01ST8CH23 WS01ST8CH26 Emiliania huxleyi Umbilicospaera sibogae WS01ST5CH10 Chrysochromulina parva WS01ST3CH23 Calcidiscus leptoporus Platychrysis sp Prymnesium parvum P994AH12 WS01ST5CH2 Unk nown deeply rooted chromophytes 99 WS01ST8CH12 WS01ST8CH15 73 70 SIP (cont.) 96 Who is there ? 0.05 Who is eating apple pie ? 65 WS01ST3SY29 WS01ST7SY24 A13 WS01ST3SY2 P99SY12 S13 GG3L P99SY5 B13 N5D P99SY1 Marine Synechococcus WS01ST2SY27 WS01ST6SY9 70 J15 WS01ST6SY3 WS01ST8SY9 WS01ST8SY18 WS01ST2SY30 70 WS01ST2SY26 95 WS01ST2SY4 P99SY22 98 Prochlorococcus marinus PAC1 WS01ST2SY19 WS01ST8SY4 WS01ST8SY26 77 Prochlorococcus marinus SB Prochlorococcus marinus GP2 91 Prochlorococcus WS01ST1SY15 83 WS01ST3SY1 WS01ST3SY5 WS01ST2SY24 WS01ST2SY33 WS01ST2SY35 Hydrogenovibrio marinus 98 WS01ST4SY12 97 WS01ST8SY15 Trichodesmium Trichodesmium thiebautii Prochlorothrix hollandica 66 WS01ST4SY3 78 WS01ST6SY8 81 WS01ST5SY21 81 Pycnococcus provasolii WS01ST3SY25 WS01ST1SY3 99 WS01ST8SY13 WS01ST8SY25 96 WS01ST8SY3 Spniach WS01ST1SY10 WS01ST8SY7 70 WS01ST5SY4 WS01ST7SY6 99 Chlamydomonas reinhardtii 74 WS01ST2SY2 WS01ST4SY17 Chlorophytes 97 WS01ST3SY26 87 WS01ST7SY29 78 WS01ST3SY4 WS01ST4SY39 81 WS01ST4SY7 Chlorella Chlorella ellipsoidea 69 Bathycoccus prasinos WS01ST1SY35 Platydorina caudata Yamagishiella unicocca WS01ST5SY7 WS01ST4SY23 92 WS01ST6SY14 67 WS01ST5SY20 WS01ST6SY26 0.05 FISH (Fluorescent In Situ Hybridization) A cell population is fixed with formaldehyde The cell membranes are permeablized DNA or RNA probe is hybridized to cells In-Situ i.e. while the cells are still mostly intact The oligonucleotide contains a fluorescent label, which can be visualized by epifluorescence microscopy FISH (cont.) Advantages Allows visualization of a particular population of cells (e.g. a species of interest) Gives quantitative information about a microbial population Can probe for DNA, mRNA and ribosomal RNA (bacterial population) (chromosome mapping) Disadvantages Cross-hybridization Different groups often do not add up to 100% of the population Relatively expensive and time consuming Microautoradiography of labeled substrate and fluorescent in situ hybridization Allows for co-localization of radiolabel and phylogenetic probe DAPI and Flo-probed cells exposed to 3H amino acids Cottrell and Kirchman 2000, AEM 66: 1692–1697 Take home messages: Molecular methods Most people prefer to work with DNA, because it is easiest and there are now many standard methods, reagents, and kits PCR based techniques have important limitations/biases DNA based methods can not determine who is active Phylogenetic analysis can not be used to calculate diversity indices (like the Shannon index) Molecular methods should be put into context of the biology and ecology of a system THE BETTER OUR METHODS THE MORE WE LEARN