notes
advertisement

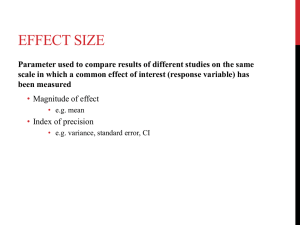
Phylogenetic comparative methods
Comparative studies (nuisance)
Evolutionary studies (objective)
Community ecology (lack of alternatives)
Current growth of phylogenetic
comparative methods
New statistical methods
Availability of phylogenies
Culture
One of many possible types of
problems
y b0 b1 x
or as a special case
y b0
This model structure can be used for
a variety of types of problems
y b0 b1 x
Assumptions:
y takes continuous values
x can be a random variable or a set of
known values (continuous or not)
y is linearly related to x
are random variables with expectation 0
and finite (co)variances that are known
y b0 b1 x
Statistical methods
(P)IC = GLS
Phylogenetic independent contrasts
Generalized Least Squares
(these are methods, not models)
Other methods for other statistical models
ML, REML, EGLS, GLM, GLMM, GEE,
“Bayesian” methods
y b0 b1 x
are random variables with expectation 0
and finite (co)variances that are known
Phylogeny provides a hypothesis for these
covariances
Close
Relatives
Tend to
Resemble
Each Other
A
B
4
C
3
D
2
E
F
Y
G
H FE
A
C
1
0
BD
G
-1
H
I
0
1
2
X
3
4
A
B
4
C
3
D
2
E
F
Y
What does this
G
represent?
H
FE
A is it
How
constructed?
C
1
0
G
-1
H
I
0
Is itDknown for
B
1 certain?
2
3
X
4
Assume that this
represents time and
is knownGwithout
errorH F E
A
B
4
C
3
D
2
E
F
G
Y
Translate
into the
C
pattern
of covariances
0
in among
species
D
1
-1
H
I
A
B
0
1
2
X
V
3
4
Trait value
Hypothetical trait for a single species
under Brownian motion evolution
possible
course of
evolution
Time
Trait value
another
possible
course of
evolution
Time
Trait value
another
possible
course of
evolution
Time
Trait value
Brownian motion evolution gives the
hypothetical variance of a trait
Variance
Time
Trait value
Brownian motion evolution
Variance
Time
Brownian motion evolution of a
hypothetical trait during speciation
Variance between
species = Time
Total variance = Total time
Variance between
species = Time
Total variance = Total time
Covariance =
Shared time
Variance between
species = Time
A
B
4
C
3
D
2
E
F
Y
G
BrownianH
motion
A
V
C
1
0
BD
G
-1
H
I
FE
0
1
2
3
4
X
V
Covariance matrix giving
phylogenetic covariances
among species
v ii diagonal elements give the total variance
for species i
v ij off-diagonal elements give covariances
between species i and species j
I am confused by the authors use of "branch
lengths" on page 3023. I'm not sure if "different
types of branch lengths" mean different
phylogenetic analyses or something else I'm not
aware of.
Digression - non-Brownian models of evolution
Ornstein-Uhlenbeck evolution
Stabilizing
selection with
strength given
by d
selection
Time
Variance between
species < Time
Total variance << Total time
Variance between
species < Time
Ornstein-Uhlenbeck evolution
Time
Variance
Stabilizing selection means information is “lost”
through time
Phylogenetic correlations between species
decrease
Phylogenetic Signal
(Blomberg, Garland, and Ives
2003)
OU
process
Vd
Vd=
measures the strength of signal
Vd=
y b0 b1 x
Assumptions:
y takes continuous values
x can be a random variable or a set of known
numbers
y is linearly related to x
are random variables with expectation 0 and
finite (co)variances that are known
If d must be estimated, cannot be analyzed using PIC
or GLS
If we are dealing with a recent, rapid radiation, (supported
clade but with short branches) will the lack of branch
length data render any PIC not very informative
biologically, because we would expect non-significant
probabilities, based solely on the branch lengths alone?
page 3022, second paragraph.
Phylogenetic Signal
(Blomberg, Garland, and Ives
2003)
OU
process
Vd
Vd=
measures the strength of signal
y b0 b1 x
Statistical methods
(P)IC = GLS
Phylogenetic independent contrasts
Generalized Least Squares
(these are methods, not models)
Other methods for other statistical models
ML, REML, EGLS, GLM, GLMM, GEE,
“Bayesian” methods
PIC
yij 1xij 'i ' j ij
'k 'l
'i i
'k 'l
4
1
y4
2
3
y1
y2
y3
4
1
y4
2
3
y1
y2
y3
y12 y1 y 2
y1 1 y 2 2 y1 y 2 1 2
y4
1 1 1 2
1 2 1 2
y 34 y 3 y 4
1 2
'4 4
1 2
PIC
yij 1xij 'i ' j ij
y ij
'i ' j
1
x ij
'i ' j
ij
Regression through the origin
PIC
y ij
'i ' j
1
x ij
'i ' j
ij
You could also use different branch lengths
for x:
y ij
'i ' j
1
x˜ ij
u'i u' j
ij
Branch lengths of y
Branch lengths of x
PIC
y ij
'i ' j
1
x ij
'i ' j
ij
You could also use different branch lengths
for x:
y ij
'i ' j
1
x˜ ij
u'i u' j
When could this be justified?
ij
When could this be justified?
y ij
'i ' j
1
x˜ ij
u'i u' j
ij
yij 1xij 'i ' j ij
Never (?)
y b0 b1 x
Statistical methods
(P)IC = GLS
Phylogenetic independent contrasts
Generalized Least Squares
(these are methods, not models)
Other methods for other statistical models
ML, REML, EGLS, GLM, GLMM, GEE,
“Bayesian” methods
y b0 b1 x
E' V I
2
2
Elements of V are given by shared
branch lengths under the
assumption of “Brownian motion”
evolution
Generalized Least Squares, GLS
y y1 ,y 2 ,...,y n
'
X 1,x
b b0 , b1
'
ˆb X' V 1X 1 X' V 1 y
'
1
ˆ
ˆ
y Xb V y X bˆ n 2
2
Ordinary least squares
ˆb X' X1 X' y
n 2
'
ˆ
ˆ y Xb y Xbˆ
2
V=I
Related to ordinary least squares
DVD' I
z Dy
U DX
y Xb
Dy DXb D
z Ub
z Ub
E' EDD '
DE' D'
D VD' I
2
2
z Ub
E ' I
2
Values of
z Dy
are linear combinations of yi
A
B
4
C
3
D
2
E
F
Y
G
H FE
A
C
1
0
BD
G
-1
H
I
0
1
2
X
3
4
GLS
parameter true value
estimate
95% confidence
LS
estimate
interval
95% confidence
interval
b0
0
2.28
[-0.82, 5.38]
-1.10
[-3.69, 1.49]
b1
0
-0.43
[-1.45, 0.60]
1.45
[0.28, 2.62]
2
2
3.35
E{Yh }
2.84
1.39
[ -0.35 , 6.03]
3.84
[0.35 , 7.33]
If IC and GLS can yield identical results and the authors
refer to IC as "a special case of GLS models" (p. 3032),
in what situation(s) would GLS be a more appropriate
method? In other words, why not just use IC?
Divergence time for desert and montane ringtail
populations assumed to be 10,000 years
QuickTime™ and a
TIFF (LZW) decompressor
are needed to see this picture.
Predicting values
for ancestral and
new species
yij 1xij 'i ' j ij
A
B
C
D
E
F
Is the prediction of the
4
estimateGof y for species I
3
H F Eprecise than
more
or less
what
you
2
A would expect from
Y a standard regression
1
C
analysis?
0
BD
G
-1
H
I
0
1
2
X
3
4
When dealing with multiple, incongruent gene trees, we can
perform multiple PIC's on each tree, and find a
correlation or not. How do we know which is the "right"
answer?
The three main phylogenetically based statistical methods
described in the reading (IC, GLS, and Monte Carlo
simulations) rely on correct information about tree
topology and branch lengths. If we are unsure of the
correctness of these basic assumptions, what is the best
way to analyze our data?
I'm unclear how data can be statistically significant when
transformed, but not significant otherwise. This seems
like cheating/lying.
The paper discussed researchers' decisions about branch
lengths, especially in terms of transformations (OU,
ACDC). Do researchers use ultrametric trees for these
analyses?