Important statistical distributions
advertisement
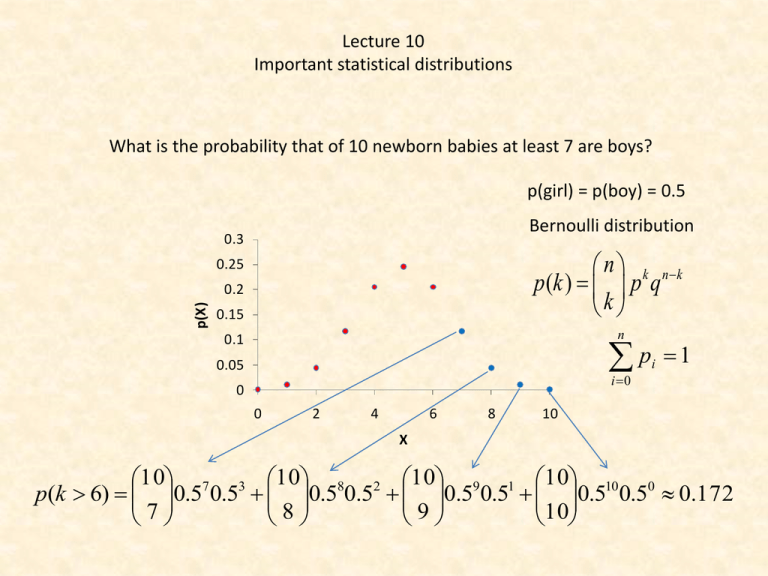
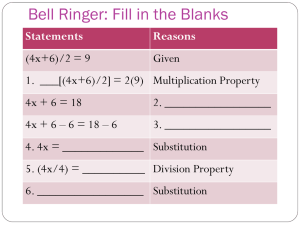
Lecture 10 Important statistical distributions What is the probability that of 10 newborn babies at least 7 are boys? p(girl) = p(boy) = 0.5 Bernoulli distribution 0.3 n k nk p (k ) p q k 0.25 p(X) 0.2 0.15 n p 0.1 0.05 i 0 0 0 2 4 6 8 i 1 10 X 10 7 3 10 8 2 10 9 1 10 10 0 p(k 6) 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.5 0.172 7 8 9 10 Bernoulli or binomial distribution n k nk p (k ) p q k 0.35 0.3 n x n x F (k ) p( x k ) p q x 0 x k np f(p) 0.25 0.2 0.15 0.1 0.05 0 0 2 npq 1 2 3 4 5 p 6 7 8 10 p(k ) 0.2k 0.810 k k The Bernoulli or binomial distribution comes from the Taylor expansion of the binomial n i n1 n n i ( p q) p q p (1 q) n1 i 0 i i 0 i n n 9 10 Assume the probability to find a certain disease in a tree population is 0.01. A biomonitoring program surveys 10 stands of trees and takes in each case a random sample of 100 trees. How large is the probability that in these stands 1, 2, 3, and more than 3 cases of this disease will occur? 1000 0.01* 0.99999 0.0004 p(1) 1 1000 0.012 * 0.99998 0.0022 p(2) 2 Mean, variance, standard deviation 1000* 0.01 10 2 1000* 0.01* 0.99 9.9 1000 0.013 * 0.99997 0.0074 p(3) 3 9.9 3.146 1000 0.010 0.991000 p(k 3) 1 p (k 3) 1 0.01 0.99 1 i 0 0 1000 1000 1000 1 999 2 998 0.01 0.99 0.01 0.99 0.0130.99997 0.99 1 2 3 3 i n i n k nk p (k ) p q k What happens if the number of trials n becomes larger and larger and p the event probability becomes smaller and smaller. np rp r p q 1 p 1 p r r (r k )! k rr k 1 (r k )! p( X k ) k !(r 1)! (r ) k ( r ) r k ! r ( r 1)!( r ) k 1 r lim r 1 e 1 r (r k )! lim r 1 k (r 1)!(r ) r p( X k ) k k! e Poisson distribution The distribution or rare events Assume the probability to find a certain disease in a tree population is 0.01. A biomonitoring program surveys 10 stands of trees and takes in each case a random sample of 100 trees. How large is the probability that in these stands 1, 2, 3, and more than 3 cases of this disease will occur? Poisson solution 1000* 0.01 10 Bernoulli solution p (1) 0.0004 10 10 e 0.00045 1! 102 10 p ( 2) e 0.0023 2! 103 10 p (3) e 0.0076 3! p (1) p (2) 0.0022 p (3) 0.0074 The probability that no infected tree will be detected 100 10 p(0) e e 10 0.000045 0! p(0) e The probability of more than three infected trees Bernoulli solution p(0) p(1) p(2) p(3) 0.00045 0.0023 0.0076 0.019 p(k 3) 1 0.019 0.981 p(k 3) 0.99 p(k) 0.4 0.35 0.3 0.25 0.2 0.15 0.1 0.05 0 =1 =2 =3 0 1 2 3 4 =4 5 =6 6 7 8 9 10 11 12 13 k 2 Variance, mean 1 Skewness What is the probability in Duży Lotek to have three times cumulation if the first time 14 000 000 people bet, the second time 20 000 000, and the third time 30 000 000? The probability to win is p ( 6) 6!43! 1 49! 14000000 1 1 14000000 1 14000000 1 2 20000000 1.428571 14000000 1 3 30000000 2.142857 14000000 The probability of at least one event: p(k 1) 1 e 10 1 p1 e 0.368 0! 1.4285710 1.428571 p2 e 0.239 0! 2.1428570 2.142857 p3 e 0.117 0! The zero term of the Poisson distribution gives the probability of no event The events are independent: p1, 2,3 0.368* 0.239* 0.117 0.01 The construction of evolutionary trees from DNA sequence data T→C T C A→G A G→C→G T G→C→A A A C G T T C A→G A G T G C C C T Probabilities of DNA substitution We assume equal substitution Single substitution probabilities. If the total probability for a substitution is p: Parallel substitution p A T Back substitution p p p Multiple substitution C G p p(A→T)+p(A→C)+p(A→G)+p(A→A) =1 The probability that A mutates to T, C, or G is P¬A=p+p+p The probability of no mutation is pA=1-3p Independent events The probability that A mutates to T and C to G is PAC=(p)x(p) Independent events p( A B) p( A) p( B) p( A B) p( A) p( B) The probability matrix T→C T C A→G A G→C→G T G→C→A A A C G T T C A→G A G T G C C C T Single substitution Parallel substitution Back substitution Multiple substitution T A C G p p p 1 3 p 1 3 p p p p P p p 1 3 p p p p p 1 3 p A T C G What is the probability that after 5 generations A did not change? p5 (1 3 p)5 The Jukes - Cantor model (JC69) now assumes that all substitution probabilities are equal. The Jukes Cantor model assumes equal substitution probabilities within these 4 nucleotides. Arrhenius model p p p 1 3 p dP (t ) P(t ) P(t ) P(0)e t p 1 3 p p p P dt p p 1 3 p p Substitution probability after time t p p p 1 3 p A,T,G,C A Transition matrix t P(t ) P(0)t The probability that nothing changes is the zero term of the Poisson distribution P( A C, T , G) e e4 pt Substitution matrix 1 3 4 t e 4 4 1 1 e 4 t P4 4 1 1 4 t 4 4e 1 1 e 4 t 4 4 1 1 4 t e 4 4 1 3 4 t e 4 4 1 1 4 t e 4 4 1 1 4 t e 4 4 1 1 4 t e 4 4 1 1 4 t e 4 4 1 3 4 t e 4 4 1 1 4 t e 4 4 The probability of at least one substitution is 1 1 4 t e 4 4 1 1 4 t e 4 4 1 1 4 t e 4 4 1 3 4 t e 4 4 P( A C T G) e 1 e4 pt The probability to reach a nucleotide from any other is 1 (1 e 4 pt ) 4 The probability that a nucleotide doesn’t change after time t is 1 1 3 P( A A, T , C , G | A) 1 3( (1 e 4 pt )) e 4 pt 4 4 4 P( A, T , G, C A) Probability for a single difference 1 3 3 P( A A, T , C , G ) 3( (1 e 4 pt )) e 4 pt 4 4 4 What is the probability of n differences after time t? 0.35 0.3 0.25 f(p) We use the principle of maximum likelihood and the Bernoulli distribution x n x n n x 3 3 3 3 p( x, t ) p (1 p) n x e4 pt 1 ( e4 pt ) 4 4 x x 4 4 0.2 0.15 0.1 0.05 0 0 1 2 3 4 5 p 6 7 8 9 n n 3 3 1 3 ln p( x, t ) ln x ln p (n x) ln(1 p) ln x ln e 4 pt (n x) ln e4 pt ) 4 4 4 4 x x t 1 4x ln1 4 p 3n This is the mean time to get x different sites from a sequence of n nucleotides. It is also a measure of distance that dependents only on the number of substitutions 10 Homo sapiens Pan troglodytes Pan paniscus Gorilla Homo neandertalensis Phylogenetic trees are the basis of any systematic classificaton t 1 4x ln1 4 p 3n Time Divergence - number of substitutions A pile model to generate the binomial. If the number of steps is very, very large the binomial becomes smooth. Abraham de Moivre (1667-1754) f ( x) Ce The normal distribution is the continous equivalent to the discrete Bernoulli distribution 1 f ( x) e 2 ( x 2 ) 1 x 2 2 -2 -1.2 -0.4 0.4 1.2 X 0.05 0.04 0.03 0.02 0.01 0 2 Frequency Frequency 0.05 0.04 0.03 0.02 0.01 0 0.06 0.04 0.02 0 -2 -1.2 -0.4 0.4 1.2 X Frequency 0.15 0.1 0.05 0 -2 -1.2 -0.4 0.4 1.2 X Frequency Frequency Frequency The central limit theorem If we have a series of random variates Xn, a new random variate Yn that is the sum of all Xn will for n→∞ be a variate that is asymptotically normally distributed. 2 2 -2 -1.2 -0.4 0.4 1.2 X 0.25 0.2 0.15 0.1 0.05 0 -2 -1.2 -0.4 0.4 1.2 X 0.15 0.1 0.05 0 -2 -1.2 -0.4 0.4 1.2 X 2 2 2 X X X 0.06 0.05 f(x) 0.04 0.03 0.02 1 f ( x) e 2 0.01 ( x )2 2 2 0 0 0.5 1 1.5 2 2.5 X 3 3.5 4 4.5 5 4 4.5 5 The normal or Gaussian distribution 1.2 1 1 F ( x) 2 f(x) 0.8 0.6 0.4 x e ( v )2 2 2 dv 0.2 0 0 0.5 1 1.5 2 2.5 X Mean: Variance: 2 3 3.5 • • Important features of the normal distribution The function is defined for every real x. The frequency at x = m is given by 1 0.4 p( x ) 2 • • The distribution is symmetrical around m. The points of inflection are given by the second derivative. Setting this to zero gives ( x ) x X X X 0.06 0.05 f(x) 0.04 - 0.03 0.02 -2 0.01 0.68 0.95 + +2 0 0 1 2 e 0.5 1 x 2 2 1 x 2 1 1 2 2 e 2 1 e 2 2 1 2 e 2 1 2 e 1 1.5 2 2.5 X 3 3.5 F ( x) 4 1 2 4.5 x e ( v )2 2 2 5 dv 0.68 1 x 2 1 x 2 2 1 x 2 2 2 0.95 0.5 0.975 Many statistical tests compare observed values with those of the standard normal distribution and assign the respective probabilities to H1. The Z-transform 1 f ( x) e 2 1 x 2 The standard normal 1 2 Z 2 f ( x) e 2 1 2 x Z The variate Z has a mean of 0 and and variance of 1. A Z-transform normalizes every statistical distribution. Tables of statistical distributions are always given as Ztransforms. The 95% confidence limit 0.1 0.04 0.06 0.04 0.02 0 0.05 0 0 2 0.02 0 0 6 12 18 24 30 36 42 48 The Z-transformed (standardized) normal distribution X X X 4 6 8 10 0 3 6 9 12 15 18 0.06 0.05 f(x) 0.04 - 0.03 0.02 -2 0.01 0.68 0.95 + +2 0 0 0.5 1 1.5 2 2.5 X 3 P( - < X < + ) = 68% P( - 1.65 < X < + 1.65) = 90% P( - 1.96 < X < + 1.96) = 95% P( - 2.58 < X < + 2.58) = 99% P( - 3.29 < X < + 3.29) = 99.9% 3.5 4 4.5 5 The Fisherian significance levels Why is the normal distribution so important? The normal distribution is often at least approximately found in nature. Many additive or multiplicative processes generate distributions of patterns that are normal. Examples are body sizes, intelligence, abundances, phylogenetic branching patterns, metabolism rates of individuals, plant and animal organ sizes, or egg numbers. Indeed following the Belgian biologist Adolphe Quetelet (17961874) the normal distribution was long hold even as a natural law. However, new studies showed that most often the normal distribution is only a approximation and that real distributions frequently follow more complicated unsymmetrical distributions, for instance skewed normals. The normal distribution follows from the binomial. Hence if we take samples out of a large population of discrete events we expect the distribution of events (their frequency) to be normally distributed. The central limit theorem holds that means of additive variables should be normally distributed. This is a generalization of the second argument. In other words the normal is the expectation when dealing with a large number of influencing variables. Gauß derived the normal distribution from the distribution of errors within his treatment of measurement errors. If we measure the same thing many times our measurements will not always give the same value. Because many factors might influence our measurement errors the central limit theorem points again to a normal distribution of errors around the mean. In the next lecture we will see that the normal distribution can be approximated by a number of other important distribution that form the basis of important statistical tests. The estimation of the population mean from a series of samples x,s x,s n xi n x n i n n x Z i 1 n i 1 n 2 si n x,s x,s x,s i 1 n=10 0.25 x,s f(x) f(x) 0.2 0.15 0.1 , 0.05 0 0 2 4 6 8 10 x,s 0.2 0.18 0.16 0.14 0.12 0.1 0.08 0.06 0.04 0.02 0 0.12 n=20 n=50 0.1 Zx n 0.08 f(x) 0.3 x,s The n samples from an additive random variate. Z is asymptotically normally distributed. 0.06 0.04 0.02 0 0 3 X 6 9 12 15 18 X 0 6 12 18 24 30 36 42 48 X 0.06 x n 0.05 f(x) 0.04 - 0.03 0.02 -2 0.01 0.68 0.95 Standard error + Confidence limit of the estimate of a mean from a series of samples. +2 0 0 0.5 1 1.5 2 2.5 X 3 3.5 4 4.5 5 is the desired probability level. How to apply the normal distribution Intelligence is approximately normally distributed with a mean of 100 (by definition) and a standard deviation of 16 (in North America). For an intelligence study we need 100 persons with an IO above 130. How many persons do we have to test to find this number if we take random samples (and do not test university students only)? 1 F ( x 130) 2 ( v )2 2 2 e 130 1 dv 1 2 a ( z ) F ( x a) 2 130 ( v ) 2 2 e dv 0.03 0.025 f(IQ) 0.02 0.015 0.01 IQ<130 IQ>130 0.005 0 40 60 80 100 IQ 120 140 160 One and two sided tests We measure blood sugar concentrations and know that our method estimates the concentration with an error of about 3%. What is the probability that our measurement deviates from the real value by more than 5%? Albinos are rare in human populations. Assume their frequency is 1 per 100000 persons. What is the probability to find 15 albinos among 1000000 persons? 1000000 15 999985 p( X 15) (0.00001) (0.99999) 15 =KOMBINACJE(1000000,15)*0.00001^15*(1-0.00001)^999985 = 0.0347 np 2 npq Home work and literature Refresh: Literature: • • • • • • • Łomnicki: Statystyka dla biologów Mendel: http://en.wikipedia.org/wiki/Mendelian_inheritan ce Pearson Chi2 test http://en.wikipedia.org/wiki/Pearson's_chisquare_test Bernoulli distribution Poisson distribution Normal distribution Central limit theorem Confidence limits One, two sided tests Z-transform Prepare to the next lecture: • • • • • • c2 test Mendel rules t-test F-test Contingency table G-test