Document
advertisement

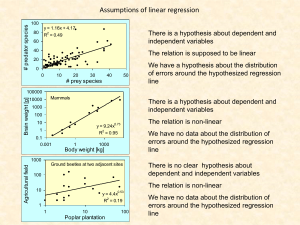
General additive models Variance and covariance a1 a2 T U U a1 ... a n a2 ... an T M M ... n UU T a 2 i Variance i 1 1 ... n a n 1 2 i 2 i 1 a1 a2 V ... a n 1 V n a n 1 i i 1 T a1 2 a2 n VV T a ... an ( n 1)Variance 2 i i 1 Sums of squares Variance 1 n 1 VV T 2 1 n 1 ( U M )( U M ) a1 A b1 B a 2 b2 B A A ; B ... ... a b A B n n n AB T a i A b i B ( n 1) Covariance i 1 1 n 1 T ( A X A ) ( B X B ) Co var iance T M contains the mean 1 n 1 ( A X A ) ( B X B ) Co var iance T The coefficient of correlation r We deal with samples cov( xy ) X Y R 1 n 1 cov( xy ) x var( y ) x y For a matrix X that contains several variables holds Cov Matrix D 1 n 1 1 n 1 ( X Μ X )' ( X Μ X ) R ( X Μ Y )' ( X Μ Y ) ( X Μ X )' ( Y Μ Y ) X xy ( X Μ X )' ( Y Μ Y ) var( x ) (X Μ y )' ( X Μ X )( Y Μ Y )' ( Y Μ Y ) 1 1 n 1 ( X Μ )' ( X Μ ) 1 n 1 Σ X ( X Μ )' ( X Μ ) Σ X 1 R Σ X DΣ X 1 1 The diagonal matrix SX contains the standard deviations as entries. X-M is called the central matrix. The matrix R is a symmetric distance matrix that contains all correlations between the variables ΣX X 1 0 0 0 0 X 2 0 0 0 ... 0 0 0 0 0 Xn R 1 1 n 1 1 Σ X ( X Μ )' ( X Μ ) Σ X R Σ X DΣ X 1 1 Pre-and postmultiplication 1 / X 1 ... X ... 1 / Xn R ΣX X T 1 / X1 ... Xn ΣX r X ΣXX T 11 21 1 / 1 ... ... 1 / n ... n1 Premultiplication n R 11 X 1; n Σ n ; n X n ;1 0 ... 1/ 2 ... ... ... 0 ... 0 Σ X ... 1 / n 0 12 ... 22 ... ... ... n2 .... ij n i 1 1 / 1 0 X ... 0 1/ ... 11 21 ... n1 j 1 i n 12 ... 22 ... ... ... n2 .... 12 ... 22 ... ... ... n2 .... 1n 2n ... nn 1n 1 / 1 2 n ... ... ... nn 1 / n Postmultiplication n r ij i 1 j 11 21 ... n1 scalar j 1 1n 2n ... nn For diagonal matrices X holds R X S X XX Σ ΣXX Linear regression European bat species and environmental correlates N=62 ln(Area) ln(Number of species) 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 10.3264 10.84344 12.40519 11.61702 8.891512 5.703782 9.068777 9.019059 10.94366 7.824046 9.132379 11.27551 10.67112 7.887209 10.71945 7.243513 12.73123 13.20664 12.78555 1.871802 11.7905 11.44094 11.54248 11.16014 12.6162 9.615805 11.07637 3.258097 0 3.218876 0.693147 2.70805 2.890372 2.995732 3.178054 2.890372 3.496508 2.197225 1.609438 3.044522 2.833213 3.526361 1.098612 2.890372 3.178054 2.639057 2.639057 2.397895 0 2.397895 3.465736 3.218876 1.609438 3.496508 3.332205 0 2.397895 3.433987 2.564949 2.772589 ln( S ) a 0 a1 ln( A ) y1 x1 1 1 y 2 x2 1 1 Y a 0 a1 ... ... ... ... 1 y x 1 n n x1 x 2 a 0 ... a 1 x n Y XA Matrix approach to linear regression X is not a square matrix, hence X-1 doesn’t exist. X ' Y X ' XA X ' X 1 X ' Y X ' X X ' XA IA A A X ' X X ' Y 1 1 The species – area relationship of European bats 3.258097 0 3.218876 0.693147 2.70805 2.890372 2.995732 3.178054 2.890372 3.496508 2.197225 1.609438 3.044522 2.833213 3.526361 1.098612 2.890372 3.178054 2.639057 2.639057 2.397895 0 2.397895 3.465736 3.218876 1.609438 3.496508 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ln(Area) 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 10.3264 10.84344 12.40519 11.61702 8.891512 5.703782 9.068777 9.019059 10.94366 7.824046 9.132379 11.27551 10.67112 7.887209 10.71945 7.243513 12.73123 13.20664 12.78555 1.871802 11.7905 X' 1 1 1 1 1 1 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 X'X 3.5 R n 1 1 y = 0.2391x + 0.1468 R² = 0.4614 3 X'Y 154.2937 1647.908 2.5 2 1.5 1 -1 (X'X) (X'Y) a0 0.146808 a1 0.239144 0.5 0 -0.5 -5 Σ X ( X Μ )' ( X Μ ) Σ X 0 5 10 15 20 ln (Area) ln S 0 . 24 ln A 0 . 15 What about the part of variance explained by our model? 1 1 1 1 1 1 1 1 10.3264 10.84344 12.40519 11.61702 8.891512 5.703782 9.068777 4 62 607.1316 607.1316 6518.161 (X'X)-1 0.183521 -0.01709 -0.01709 0.001746 ln(# species) ln(Number of Constant species) S e 1 0 . 15 A 0 . 24 1 . 16 A 0 . 24 1.16: Average number of species per unit area (species density) 0.24: spatial species turnover R 0.769488 -2.48861 0.730267 -1.79546 0.219442 0.401763 0.507124 0.689445 0.401763 1.007899 -0.29138 -0.87917 0.555914 0.344605 1.037752 -1.39 0.401763 0.689445 0.150449 0.150449 -0.09071 -2.48861 -0.09071 0.977127 0.730267 -0.87917 1.007899 0.843596 -2.48861 -0.09071 0.945379 0.076341 1 n 1 Σ X ( X Μ )' ( X Μ ) Σ X 1 (X-M)' 0.473878 -3.64398 1.54459 -2.09623 -1.27246 2.451164 0.533954 1.050991 2.612741 1.824579 -0.90093 -4.08866 -0.72367 -0.77339 1.151213 -1.9684 -0.66007 1.48306 0.878671 -1.90524 0.927004 -2.54893 2.938785 3.414195 2.993105 -7.92064 1.998051 1.64849 1.750039 1.367698 2.823752 -0.17664 0.769488 0.473878 -2.48861 0.730267 -3.64398 1.54459 (X-M)'(X-M) 71.0087 136.9954 136.9954 572.8582 -1.79546 0.219442 0.401763 -2.09623 -1.27246 2.451164 (X-M)'(X-M) / (n-1) 1.164077 2.245826 2.245826 9.391119 Sx 1.078924 0 0 3.064493 4 3.5 Sx -1 0.926849 0 0 0.326318 Sx -1 (X-M)'(X-M) / (n-1) 1.078924 2.081542 0.732854 3.064493 Sx-1 (X-M)'(X-M) / (n-1) Sx-1 1 0.679245 0.679245 1 Sx-1 (X-M)'(X-M) / (n-1) Sx-1)2 1 0.461374 0.461374 1 y = 0.2391x + 0.1468 R² = 0.4614 3 ln(# species) X-M 1 2.5 2 1.5 1 0.5 0 -0.5 -5 0 5 10 ln (Area) 15 20 How to interpret the coefficient of determination 4 3.5 y = 0.2391x + 0.1468 R² = 0.4614 3 ln(# species) 2 Y ;M n 1 (Y n 1 2 Total variance 2 1.5 1 2 Y ;Y ( X ) n 1 n 1 (Y i Y ( X i )) 2 i 1 Rest (unexplained) variance 0.5 0 -0.5 2 Y ( X ); M n 1 (Y ( X n 1 i )Y) i 1 -5 0 5 10 15 20 Residual (explained) variance ln (Area) Y ; M Y ;Y ( X ) Y ( X ); M 2 n 1 2 Y) i 1 2.5 R 1 i Residual variance 1 (Y n 1 Total variance n i Y ( X i )) i 1 n 1 (Y n 1 2 i Y) Statistical testing is done by an F or a t-test. F 1 R 2 )Y) n i Y) i 1 t 2 i i 1 (Y 2 i 1 R (Y ( X df t F R 1 R 2 df 2 2 2 2 2 ln( S ) a 0 a1 ln( A ) a 2 T a 3 N T 0 a 4 L The general linear model n Y a 0 a1 X 1 a 2 X 2 a 3 X 3 ... a n X n a 0 a i Xi i 1 A model that assumes that a dependent variable Y can be expressed by a linear combination of predictor variables X is called a linear model. y1 y2 Y ... y m 1 1 1 1 y1 y2 Y ... y m 1 1 1 1 x 1 ,1 ... x 2 ,1 ... ... ... x m ,1 ... x 1 ,1 ... x 2 ,1 ... ... ... x m ,1 ... x1 , n a 0 x 2 , n y1 XA ... ... x m , n y n X ' Y X ' XA X ' X 1 X ' Y A X ' X X ' Y x1 , n a 0 0 x 2 , n y1 1 XA Ε ... ... ... x m , n y n n The vector E contains the error terms of each regression. Aim is to minimize E. X ' X X ' XA IA A 1 1 The general linear model n Y a 0 a1 X 1 a 2 X 2 a 3 X 3 ... a n X n a 0 a i Xi i 1 If the errors of the preictor variables are Gaussian the error term e should also be Gaussian and means and variances are additive n Y a 0 a1 X 1 a 2 X 2 a 3 X 3 ... a n X n a 0 a i Xi i 1 n (Y ) a 0 a i ( X i ) ( ) i 1 (Y ) a 0 2 2 Total variance 2 i 1 2 a i X i ( ) Explained variance a0 2 R n Unexplained (rest) variance a X i i 2 (Y ) 2 ( ) i 1 2 2 (Y ) (Y ) n ln( S ) a 0 a1 ln( A ) a 3 N T 0 a 4 L Y Country/Island Albania Andorra Austria Azores Baleary Islands Belarus Belgium Bosnia and Herzegovina British islands Bulgaria Canary Islands Channel Is. Corsica Crete Croatia Cyclades Is. Cyprus Czech Republic Denmark Dodecanese Is. Estonia Faroe Is. Finland France Germany Gibraltar Greece Hungary Iceland X ln(Number of Constant species) 3.258097 0 3.218876 0.693147 2.70805 2.890372 2.995732 3.178054 2.890372 3.496508 2.197225 1.609438 3.044522 2.833213 3.526361 1.098612 2.890372 3.178054 2.639057 2.639057 2.397895 0 2.397895 3.465736 3.218876 1.609438 3.496508 3.332205 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ln(Area) Days below zero Latitude of capitals (decimal degrees) 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 10.3264 10.84344 12.40519 11.61702 8.891512 5.703782 9.068777 9.019059 10.94366 7.824046 9.132379 11.27551 10.67112 7.887209 10.71945 7.243513 12.73123 13.20664 12.78555 1.871802 11.7905 11.44094 11.54248 34 60 92 1 18 144 50 114 64 102 1 12 11 1 114 1 2 119 85 2 143 35 169 50 97 0 2 100 133 41.33 42.5 48.12 37.73 39.55 53.87 50.9 43.82 51.15 42.65 27.93 49.22 41.92 35.33 45.82 37.1 35.15 50.1 55.63 36.4 59.35 62 60.32 48.73 52.38 36.1 37.9 47.43 64.13 Multiple regression 1. Model formulation 2. Estimation of model parameters 3. Estimation of statistical significance Y XA A X ' X X ' Y 1 X' 1 1 1 1 1 1 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 144 18 1 92 60 34 53.87 39.55 37.73 48.12 42.5 41.33 1 1 10.3264 10.84344 114 50 43.82 50.9 X'X 62 607.1316 4328 2906.4 2906.4 4328 607.1316 6518.161 48545.59 29086.57 534136 228951.7 48545.59 29086.57 228951.7 141148.1 (X'X)-1 1.019166 -0.02275 -0.02275 0.002458 0.00261 -7.5E-05 -0.02053 8.3E-05 0.00261 -0.02053 -7.5E-05 8.3E-05 1.3E-05 -5.9E-05 -5.9E-05 0.000509 (X'X)-1X' 0.025783 0.163309 0.013407 0.003376 -0.00859 0.002243 -0.00017 0.000405 9.87E-05 -0.00066 -0.00195 -0.00056 a0 a1 a2 a3 (X'X)-1X'Y 2.679757 0.290121 0.002155 -0.06789 0.07203 0.060295 0.010457 -0.13031 0.170347 -0.00078 0.00013 0.001069 0.003124 -0.00097 -0.00019 -0.00014 0.000364 -0.00054 0.000676 -0.00074 -0.00076 -0.00064 0.003269 -0.00409 X'Y 154.2937 1647.908 11289.32 7137.716 (X'X)-1(X'Y) 2.679757 0.290121 0.002155 -0.06789 Multiple R and R2 The coefficient of determination n 1 R 1 2 Residual variance Total variance 1 (Y n 1 n i Y ( X i )) i 1 1 n (Y n 1 2 i Y) (Y ( X i 1 R 1 1 n 1 Σ X ( X Μ )' ( X Μ ) Σ X y x1 x2 1 r1 y R r2 y ... r2 y ry 1 ry 2 ... 1 r21 ... r21 ... ... ... ... ... rn 1 ... ... 1 R R XY )Y) 2 i 1 n (Y 2 i i Y) 2 i 1 1 xm rmy rm 1 ... ... 1 R YX R XX The correlation matrix can be devided into four compartments. T R R XY R XX 2 R 2 1 R YX R XY R XX det( R XX ) det( R ) det( R XX ) 1 1 R XY T det( R ) det( R XX ) ln(Number of species) ln(Area) 3.2580965 0 3.2188758 0.6931472 2.7080502 2.8903718 2.9957323 3.1780538 2.8903718 3.4965076 2.1972246 1.6094379 3.0445224 2.8332133 3.5263605 1.0986123 2.8903718 3.1780538 2.6390573 2.6390573 2.3978953 0 2.3978953 3.4657359 3.2188758 1.6094379 3.4965076 3.3322045 0 2.3978953 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 10.3264 10.84344 12.40519 11.61702 8.891512 5.703782 9.068777 9.019059 10.94366 7.824046 9.132379 11.27551 10.67112 7.887209 10.71945 7.243513 12.73123 13.20664 12.78555 1.871802 11.7905 11.44094 11.54248 11.16014 3.4339872 2.5649494 2.7725887 1.7917595 2.6390573 2.8903718 12.6162 9.615805 11.07637 5.075174 11.08702 7.858641 Days below zero 34 60 92 1 18 144 50 114 64 102 1 12 11 1 114 1 2 119 85 2 143 35 169 50 97 0 2 100 133 23 18 Latitude of capitals (decimal degrees) 41.33 42.5 48.12 37.73 39.55 53.87 50.9 43.82 51.15 42.65 27.93 49.22 41.92 35.33 45.82 37.1 35.15 50.1 55.63 36.4 59.35 62 60.32 48.73 52.38 36.1 37.9 47.43 64.13 53.43 41.8 110 124 90 130 93 52.7 56.96 47.67 54.62 49.62 X-M X-M X-M X-M (X-M)' 0.769488 -2.48861 0.730267 -1.79546 0.219442 0.401763 0.507124 0.689445 0.401763 1.007899 -0.29138 -0.87917 0.555914 0.344605 1.037752 -1.39 0.401763 0.689445 0.150449 0.150449 -0.09071 -2.48861 -0.09071 0.977127 0.730267 -0.87917 1.007899 0.843596 -2.48861 -0.09071 0.473878 -3.64398 1.54459 -2.09623 -1.27246 2.451164 0.533954 1.050991 2.612741 1.824579 -0.90093 -4.08866 -0.72367 -0.77339 1.151213 -1.9684 -0.66007 1.48306 0.878671 -1.90524 0.927004 -2.54893 2.938785 3.414195 2.993105 -7.92064 1.998051 1.64849 1.750039 1.367698 -35.8065 -9.80645 22.19355 -68.8065 -51.8065 74.19355 -19.8065 44.19355 -5.80645 32.19355 -68.8065 -57.8065 -58.8065 -68.8065 44.19355 -68.8065 -67.8065 49.19355 15.19355 -67.8065 73.19355 -34.8065 99.19355 -19.8065 27.19355 -69.8065 -67.8065 30.19355 63.19355 -46.8065 -5.54742 -4.37742 1.242581 -9.14742 -7.32742 6.992581 4.022581 -3.05742 4.272581 -4.22742 -18.9474 2.342581 -4.95742 -11.5474 -1.05742 -9.77742 -11.7274 3.222581 8.752581 -10.4774 12.47258 15.12258 13.44258 1.852581 5.502581 -10.7774 -8.97742 0.552581 17.25258 6.552581 0.769488 0.473878 -35.8065 -5.54742 (X-M)'(X-M) 71.0087 136.9954 518.6241 -95.1758 -2.48861 0.730267 -3.64398 1.54459 -9.80645 22.19355 -4.37742 1.242581 136.9954 572.8582 6163.884 625.8081 -1.79546 0.219442 0.401763 0.507124 0.689445 0.401763 1.007899 -2.09623 -1.27246 2.451164 0.533954 1.050991 2.612741 1.824579 -68.8065 -51.8065 74.19355 -19.8065 44.19355 -5.80645 32.19355 -9.14742 -7.32742 6.992581 4.022581 -3.05742 4.272581 -4.22742 518.6241 -95.1758 6163.884 625.8081 232013.7 26066.26 26066.26 4903.6 (X-M)'(X-M)/(n-1) 1.164077 2.245826 2.245826 9.391119 8.502034 101.0473 -1.56026 10.25915 2 T 1 S1 0.926849 0 0 0 0 0.326318 0 0 0 0 0.016215 0 0 0 0 0.111534 S1 D 1.078924 0.732854 0.137858 -0.17402 2.081542 3.064493 1.638448 1.144244 7.880104 -1.44613 32.97357 3.347747 61.67255 6.928784 47.66024 8.965874 R 2 det( R XX ) det( R ) det( R XX ) S1 DS-1 0.945379 2.823752 -51.8065 -5.07742 0.076341 -0.17664 40.19355 5.822581 0.28398 1.283927 54.19355 10.08258 -0.69685 -4.71727 20.19355 0.792581 0.150449 1.294578 60.19355 7.742581 0.401763 -1.9338 23.19355 2.742581 R YX R XY R XX 1 R XY 8.502034 -1.56026 101.0473 10.25915 3803.503 427.3157 427.3157 80.38689 S 1.078924 0 0 0 0 3.064493 0 0 0 0 61.67255 0 0 0 0 8.965874 1 0.679245 0.127773 -0.16129 0.679245 1 0.534656 0.373388 0.127773 0.534656 1 0.772795 -0.16129 0.373388 0.772795 1 Det RXX 0.286065 Det R 0.095413 T S1 DS-1 )-1 1.408029 -0.86031 0.139099 -0.86031 3.008345 -2.00361 0.139099 -2.00361 2.496439 S1 DS-1 )-1 RXY 0.824037 0.123194 -0.56418 S1 DS-1 )-1 RXYRYX 0.666462 1 det( R ) det( R XX ) R2 0.666462 S1 DS-1 1 0.679245 0.127773 -0.16129 0.679245 1 0.534656 0.373388 0.127773 0.534656 1 0.772795 -0.16129 0.373388 0.772795 1 R R XY R XX -0.29138 -0.90093 -68.8065 -18.9474 Det RXX 0.286065 Det R 0.095413 R2 0.666462 1 SE trace ( R )( 1 R ) 2 n k 1 R: correlation matrix n: number of cases k: number of independent variables in the model t parameter SE ( parameter ) D<0 is statistically not significant and should be eliminated from the model. Adjusted R2 2 R adj 1 (1 R ) 2 1 df 1 2 n 1 n k 1 F 2 2 df 2 R n k 1 2 1 R 2 k 0 . 66646 62 3 1 0 . 33354 3 38 . 6307 A mixed model ln S a 0 a1 ln A a 2 D T 0 a 3 L a 4 L Y Country/Island Albania Andorra Austria Azores Baleary Islands Belarus Belgium Bosnia and Herzegovina British islands Bulgaria Canary Islands Channel Is. Corsica Crete Croatia Cyclades Is. Cyprus Czech Republic Denmark Dodecanese Is. Estonia Faroe Is. Finland France Germany Gibraltar Greece Hungary Iceland Ireland Italy Kaliningrad Region Latvia X ln(Number of Constant species) 3.258097 0 3.218876 0.693147 2.70805 2.890372 2.995732 3.178054 2.890372 3.496508 2.197225 1.609438 3.044522 2.833213 3.526361 1.098612 2.890372 3.178054 2.639057 2.639057 2.397895 0 2.397895 3.465736 3.218876 1.609438 3.496508 3.332205 0 2.397895 3.433987 2.564949 2.772589 2 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 ln(Area) Days below zero 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 10.3264 10.84344 12.40519 11.61702 8.891512 5.703782 9.068777 9.019059 10.94366 7.824046 9.132379 11.27551 10.67112 7.887209 10.71945 7.243513 12.73123 13.20664 12.78555 1.871802 11.7905 11.44094 11.54248 11.16014 12.6162 9.615805 11.07637 34 60 92 1 18 144 50 114 64 102 1 12 11 1 114 1 2 119 85 2 143 35 169 50 97 0 2 100 133 23 18 110 124 Latitude of capitals Latitude2 (decimal degrees) 41.33 42.5 48.12 37.73 39.55 53.87 50.9 43.82 51.15 42.65 27.93 49.22 41.92 35.33 45.82 37.1 35.15 50.1 55.63 36.4 59.35 62 60.32 48.73 52.38 36.1 37.9 47.43 64.13 53.43 41.8 52.7 56.96 1708.169 1806.25 2315.534 1423.553 1564.203 2901.977 2590.81 1920.192 2616.323 1819.023 780.0849 2422.608 1757.286 1248.209 2099.472 1376.41 1235.523 2510.01 3094.697 1324.96 3522.423 3844 3638.502 2374.613 2743.664 1303.21 1436.41 2249.605 4112.657 2854.765 1747.24 2777.29 3244.442 X' 1 1 1 1 1 1 10.26632 6.148468 11.33704 7.696213 8.519989 12.24361 34 60 92 1 18 144 41.33 42.5 48.12 37.73 39.55 53.87 1708.169 1806.25 2315.534 1423.553 1564.203 2901.977 1 1 10.3264 10.84344 50 114 50.9 43.82 2590.81 1920.192 X'X 62 607.1316 4328 2906.4 141148.1 607.1316 4328 2906.4 141148.1 6518.161 48545.59 29086.57 1441737 48545.59 534136 228951.7 12488619 29086.57 228951.7 141148.1 7106497 1441737 12488619 7106497 3.71E+08 (X'X)-1 6.45421 0.000497 0.001087 -0.25606 0.002409 0.000497 0.002557 -8.1E-05 -0.00092 1.03E-05 0.001087 -8.1E-05 1.34E-05 6.63E-06 -6.8E-07 -0.25606 -0.00092 6.63E-06 0.010716 -0.0001 0.002409 1.03E-05 -6.8E-07 -0.0001 1.07E-06 (X'X)-1X' 0.028519 -0.00857 -0.18332 0.227512 0.119213 -0.18587 -0.27812 -0.01106 0.003388 -0.00932 0.001402 -0.00011 0.000382 0.000229 0.002492 -0.00174 -0.00017 0.000453 0.000154 -0.00024 -0.00016 0.000419 -0.00049 0.000727 -0.00078 0.0055 0.007968 -0.00748 -0.00331 0.007864 0.009674 0.003767 1.21E-06 -7.6E-05 -8.7E-05 6.89E-05 2.61E-05 -8.7E-05 -6.6E-05 -8E-05 a0 a1 a2 a3 a4 (X'X)-1X'Y -3.40816 0.264082 0.003862 0.195932 -0.0027 The final model ln S 3 . 41 0 . 26 ln A 0 . 004 D T 0 0 . 196 L 0 . 0027 L 2 Negative species density Realistic increase of species richness with area Increase of species richness with winter length Increase of species richness at higher latitudes A peak of species richness at intermediate latitudes ln(# species predicted) Is this model realistic? The model makes a series of unrealistic predictions. Our initial assumptions are wrong despite of the high degree of variance explanation 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 -0.5 -1 0 y = 0.6966x + 0.7481 R² = 0.6973 1 2 3 4 Our problem arises in part from the intercorrelation between the predictor variables (multicollinearity). We solve the problem by a stepwise approach eliminating the variables that are either not significant or give unreasonable parameter values ln (# species observed) The variance explanation of this final model is higher than that of the previous one. Multiple regression solves systems of intrinsically linear algebraic equations 2 3 2 3 Y a 10 a 11 X 1 a 12 X 1 a 13 X 1 ... a 21 X 2 b 22 X 2 a 23 X 2 ... a n 1 X a n 2 X 2 a n 3 X ... 3 A X ' X X ' Y 1 Polynomial regression • • • • • The matrix X’X must not be singular. It est, the variables have to be independent. Otherwise we speak of multicollinearity. Collinearity of r<0.7 are in most cases tolerable. Multiple regression to be safely applied needs at least 10 times the number of cases than variables in the model. Statistical inference assumes that errors have a normal distribution around the mean. The model assumes linear (or algebraic) dependencies. Check first for non-linearities. Check the distribution of residuals Yexp-Yobs. This distribution should be random. Check the parameters whether they have realistic values. Multiple regression is a hypothesis testing and not a hypothesis generating technique!! ln(# species predicted) • General additive model 4.5 4 3.5 3 2.5 2 1.5 1 0.5 0 -0.5 -1 0 y = 0.6966x + 0.7481 R² = 0.6973 1 2 ln (# species observed) 3 4 Standardized coefficients of correlation Z-tranformed distributions have a mean of 0 an a standard deviation of 1. Z x B Z X ' Z X 1 Z X ' Z Y n r Z'Z 1 (X i X )(Y i Y ) i 1 n 1 Zx 1i Zx i1 sXsY ... ... ... ... ... ... ... ... ... ... Zx ni Zx i1 n 1 Zx n 1 i 1 (X i X ) (Y i Y ) sX sY n 1 n 1 Z X ZY i 1 Zx 1 n r11 ... 1 ... R Z ' Z ... ... n 1 r Zx ii Zx ii n1 ni B R xx R .... ... ... ... ... ... ... ... r1 n ... ... rnn 1 R 1 n 1 1 Σ X ( X Μ )' ( X Μ ) Σ X 1 R 1 n 1 Z'Z XY R XY R XX B In the case of bivariate regression Y = aX+b, Rxx = 1. Hence B=RXY. Hence the use of Z-transformed values results in standardized correlations coefficients, termed b-values then b Xi B Xi If Y BX Xi How to interpret beta-values Y Beta values are generalisations of simple coefficients of correlation. However, there is an important difference. The higher the correlation between two or more predicator variables (multicollinearity) is, the less will r depend on the correlation between X and Y. Hence other variables might have more and more influence on r and b. For high levels of multicollinearity it might therefore become more and more difficult to interpret beta-values in terms of correlations. Because beta-values are standardized b-values they should allow comparisons to be make about the relative influence of predicator variables. High levels of multicollinearity might let to misinterpretations. Beta values above one are always a sign of too high multicollinearity Hence high levels of multicollinearity might reduce the exactness of beta-weight estimates change the probabilities of making type I and type II errors make it more difficult to interpret beta-values. We might apply an additional parameter, the so-called coefficient of structure. The coefficient of structure ci is defined as ci riY R 2 where riY denotes the simple correlation between predicator variable i and the dependent variable Y and R2 the coefficient of determination of the multiple regression. Coefficients of structure measure therefore the fraction of total variability a given predictor variable explains. Again, the interpretation of ci is not always unequivocal at high levels of multicollinearity. Partial correlations 2 r zy Y 1.5 1 X 1 0.5 0.5 0 y = 1.70Z + 0.60 0 0 Y 2 Y Z r xy y = 1.02Z + 0.41 1.5 X X r zx 2.5 0.5 1 0 0.5 Z X X (Y ) X ( Z ) Y Y (X ) Y ( Z ) 1 Z The partial correlation rxy/z is the correlation of the residuals X and Y rX Y / Z rX Y rX Z rYZ 1 rX Z 2 1 rYZ 2 Semipartial correlation r( X |Y ) Z rX Y rX Z rY Z 1 rY Z 2 A semipartial correlation correlates a variable with one residual only. Path analysis and linear structure models Multiple regression X1 e X2 X3 X4 Y e The error term e contain the part of the variance in Y that is not explained by the model. These errors are called residuals Y a 0 a1 X 1 a 2 X 2 a 3 X 3 a 4 X 4 e e e X3 Y X2 X1 e X4 e Regression analysis does not study the relationships between the predictor variables Path analysis defines a whole model and tries to separate correlations into direct and indirect effects Path analysis tries to do something that is logically impossible, to derive causal relationships from sets of observations. Path analysis is largely based on the computation of partial coefficients of correlation. Y p XY W p ZY X p XW e e Z Path coefficients e p ZX e Path analysis is a model confirmatory tool. It should not be used to generate models or even to seek for models that fit the data set. X p xy Y e Z p zx X p zy Y e W p xw X e We start from regression functions p xw X W e 0 X p xy Y e 0 p zx X p zy Y Z e 0 From Z-transformed values we get p xw X W e 0 Z W p xw Z X e X p xy Y e 0 Z X p xy Z Y e p zx X p zy Y Z e 0 Z Z p zx Z X p zy Z Y e X Z W Z Y p xw Z X Z Y e Z Y pYX Y Z X Z W p xy Z Y Z W e Z W Z Z Z W p zx Z X Z W p zy Z Y Z W e Z W p XW Z X Z Z p xy Z Y Z X e Z X W p XZ Z X Z Y p xy Z Y Z Y e Z Y p YZ Z Z Z Y p zx Z X Z Y p zy Z Y Z Y e Z Y Z r W Y p x w rX Y rX W p x y rY W Path analysis is a nice tool to generate hypotheses. It fails at low coefficients of correlation and circular model structures. rZ W p z x rX W p z y rY W eZY = 0 ZYZY = 1 rX Z p x y rY X rX Y p x y rZ Y p z x rX Y p z y ZXZY = rXY Non-metric multiple regression Target symptom X 1 1 0 1 0 1 0 1 1 0 1 1 1 1 0 0 1 1 0 Predicted value Sum A 0 0 1 0 1 0 1 0 1 1 0 0 0 0 1 1 0 0 1 B 1 1 0 1 0 1 0 1 1 1 0 0 0 1 1 1 0 0 1 Symptoms C 1 1 0 1 0 1 0 1 1 0 1 1 0 0 0 0 1 1 0 D 0 0 0 1 0 1 0 1 1 0 1 1 1 1 0 0 1 1 1 E 1 1 0 1 1 1 0 1 1 1 1 1 1 1 0 0 1 1 1 8 11 10 11 15 Expected values 0.848615 0.848615 -0.2092 1.108631 0.106749 1.108631 -0.2092 1.108631 0.899435 0.19602 1.01936 1.01936 0.575961 0.665233 -0.11992 -0.11992 1.01936 1.01936 0.456037 X' A B C D E 0 1 1 0 1 X'X A A B C D E 0 1 1 0 1 B 8 5 1 2 4 1 0 0 0 0 C 5 11 6 6 9 D 1 6 10 8 10 X'Y 1 1 7 10 10 12 0.5 0 -0.5 0 1 Observed occurrences 1 0 0 0 1 E 2 6 8 11 11 4 9 10 11 15 (X'X)-1 0.205969 -0.09304 0.098145 0.0242 -0.08228 -0.09304 0.224792 -0.05216 0.028233 -0.09599 0.098145 -0.05216 0.361387 -0.06158 -0.19064 0.0242 0.028233 -0.06158 0.368379 -0.25249 -0.08228 -0.09599 -0.19064 -0.25249 0.458457 +L$25*B23+L$26*C23+L$27*D23+L$28*E23+L$29*F23 1.5 0 1 1 1 1 (X'X)-1X'Y -0.2092 0.089271 0.443399 0.260016 0.315945 Statistical inference n 1 R 2 Residual variance 1 (Y n 1 i 1 1 Total variance n 1 Predicted value i Y ( X i )) n (Y i Y) i 1 1.5 1 0.5 0 -0.5 0 1 2 2 Symptoms Target symptom X A B C D E 1 0 1 1 0 1 1 0 1 1 0 1 0 1 0 0 0 0 1 0 1 1 1 1 0 1 0 0 0 1 1 0 1 1 1 1 0 1 0 0 0 0 1 0 1 1 1 1 1 1 1 1 1 1 0 1 1 0 0 1 1 0 0 1 1 1 1 0 0 1 1 1 1 0 0 0 1 1 1 0 1 0 1 1 0 1 1 0 0 0 0 1 1 0 0 0 1 0 0 1 1 1 1 0 0 1 1 1 0 1 1 0 1 1 Mean 0.631579 0.421053 0.578947 0.526316 0.578947 0.789474 Predicted Predicted Total Explained values values variance variance Unexplain ed variance 0.848615 0.848615 -0.2092 1.108631 0.106749 1.108631 -0.2092 1.108631 0.899435 0.19602 1.01936 1.01936 0.575961 0.665233 -0.11992 -0.11992 1.01936 1.01936 0.456037 0.022917 0.022917 0.043763 0.011801 0.011395 0.011801 0.043763 0.011801 0.010113 0.038424 0.000375 0.000375 0.179809 0.112069 0.014382 0.014382 0.000375 0.000375 0.207969 1 SE n k 1 2 0.828365 R2 (X'X)-1 0.828365 0.205969 -0.09304 0.098145 0.0242 -0.08228 2 1-R -0.09304 0.224792 -0.05216 0.028233 -0.09599 0.171635 0.098145 -0.05216 0.361387 -0.06158 -0.19064 N df 0.0242 0.028233 -0.06158 0.368379 -0.25249 19 13 -0.08228 -0.09599 -0.19064 -0.25249 0.458457 B 1 0.047105 0.047105 0.706903 0.227579 0.275446 0.227579 0.706903 0.227579 0.071747 0.189711 0.150374 0.150374 0.003093 0.001133 0.564758 0.564758 0.150374 0.150374 0.030815 0.245614 0.249651 0.042156 Approximated R2 trace ( R )( 1 R ) 0.135734 0.135734 0.398892 0.135734 0.398892 0.135734 0.398892 0.135734 0.135734 0.398892 0.135734 0.135734 0.135734 0.135734 0.398892 0.398892 0.135734 0.135734 0.398892 True R2 Observed occurrences Rounding errors due to different precisions cause the residual variance to be larger than the total variance. 1 1 0 1 0 1 0 1 1 0 1 1 1 1 0 0 1 1 0 A B C D E -0.2092 0.089271 0.443399 0.260016 0.315945 0.035351 0.038582 0.062027 0.063227 0.078687 SE(B) 0.052147 0.054478 0.069074 0.069739 0.0778 t -4.01163 1.638668 6.419143 3.728397 4.060988 P 0.001479 0.125246 2.27E-05 0.002529 0.001348 Logistic and other regression techniques M a le M a le M a le M a le M a le M a le M a le M a le M a le M a le F e m a le F e m a le F e m a le F e m a le F e m a le F e m a le F e m a le F e m a le F e m a le F e m a le A 5 .9 9 8 3 .9 1 6 4 .5 1 1 5 .9 4 0 6 .5 3 2 6 .5 1 3 3 .0 5 2 3 .5 1 2 6 .6 7 6 6 .9 7 6 5 .6 4 9 5 .7 1 2 5 .1 1 2 3 .6 8 1 5 .2 3 9 5 .1 8 0 2 .1 3 3 5 .3 6 1 6 .4 6 0 6 .8 3 9 B 0 .8 3 8 0 .9 9 2 0 .9 0 4 0 .7 9 5 0 .5 7 4 1 .0 3 6 0 .5 8 4 1 .1 2 6 0 .9 9 2 0 .5 0 2 0 .9 1 3 0 .4 7 4 0 .2 7 7 0 .3 2 9 0 .9 2 2 0 .5 4 6 0 .3 0 0 0 .4 7 2 0 .3 2 1 0 .4 2 6 C 2 .2 5 3 1 .9 6 4 1 .9 3 0 1 .1 7 1 1 .3 9 0 0 .5 7 1 2 .1 7 9 1 .8 4 3 2 .2 8 8 1 .0 6 2 2 .2 3 1 2 .2 3 7 1 .0 0 9 2 .4 2 0 1 .5 9 2 2 .4 1 8 3 .0 8 7 2 .1 7 5 1 .0 0 7 3 .1 7 9 Z e y 1 e y 1 1 e n Y a0 a i xi i 1 We use odds n a0 n p ln a0 1 p n ax i i 1 i p 1 p e a0 aixi i 1 p e n a0 Threshold 1 Z 0 .8 0 .6 Indecisive 0 .4 region e a i xi i 1 n 1 e a0 a i xi i 1 The logistic regression model 0 .2 0 0 5 S urely m ales 10 Y 15 S urely fem ales 20 aixi i 1 n 1 e 1 .2 Z y a0 aixi i 1 Z e 0.19 0.2 A 6.36 B 1.77 C 1 e 0.19 0.2 A 6.36 B 1.77 C 1 0 .6 0 .4 0 .2 S ex F e m a le F e m a le F e m a le F e m a le F e m a le M a le M a le M a le M a le 0 M a le Z 0 .8 Generalized non-linear regression models Y b0 1 b 1e a0 1 Y Y 1 aixi b 1= 3 b 1= 1 b 2= 4 b 2 =0 .5 0 0 0 5 x 10 0 5 10 x A special regression model that is used in pharmacology Y b0 b0 X 1 b 1 b2 b0 is the maximum response at dose saturation. b1 is the concentration that produces a half maximum response. b2 determines the slope of the function, that means it is a measure how fast the response increases with increasing drug dose.