B-cell epitopes
advertisement
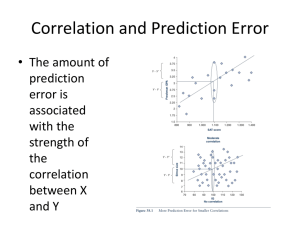
Prediction of B cell epitopes Ole Lund Outline • What is a B-cell epitope? • How can you predict B-cell epitopes? What is a B-cell epitope? B-cell epitopes Antibody Fab fragment Accessible structural feature of a pathogen molecule. Antibodies are developed to bind the epitope specifically using the complementary determining regions (CDRs). B-cell epitope classification B-cell epitope – structural feature of a molecule or pathogen, accessible and recognizable by B-cells Linear epitopes One segment of the amino acid chain Discontinuous epitope (with linear determinant) Discontinuous epitope Several small segments brought into proximity by the protein fold The Antibody • Two light and heavy chains • High variability in the complementary determine regions (CDR) • ~2.5 * 107 different phenotypes Binding of a discontinuous epitope Antibody FAB fragment complexed with Guinea Fowl Lysozyme (1FBI). Black: Light chain, Blue: Heavy chain, Yellow: Residues with atoms distanced < 5Å from FAB antibody fragments. Guinea Fowl Lysozyme KVFGRCELAAAMKRHGLDNYRGYSLGNWVCAAKFESNFNSQNRNTDGS DYGVLNSRWWCNDGRTPGSRNLCNIPCSALQSSDITATANCAKKIVSDG GMNAWVAWRKCKGTDVRVWIKGCRL B-cell epitope annotation • Linear epitopes: – Chop sequence into small pieces and measure binding to antibody • Discontinuous epitopes: – Measure binding of whole protein to antibody • The best annotation method : X-ray crystal structure of the antibody-epitope complex B-cell epitope data bases • Databases: IEDB, AntiJen, BciPep, Los Alamos HIV database, Protein Data Bank • Large amount of data available for linear epitopes • Few data available for discontinuous epitopes Sequence-based methods for prediction of linear epitopes Protein hydrophobicity – hydrophilicity algorithms Parker, Fauchere, Janin, Kyte and Doolittle, Manavalan Sweet and Eisenberg, Goldman, Engelman and Steitz (GES), von Heijne Protein flexibility prediction algorithm Karplus and Schulz Protein secondary structure prediction algorithms GOR II method (Garnier and Robson), Chou and Fasman, Pellequer Protein “antigenicity” prediction : Hopp and Woods, Welling TSQDLSVFPLASCCKDNIASTSVTLGCLVTG YLPMSTTVTWDTGSLNKNVTTFPTTFHETY GLHSIVSQVTASGKWAKQRFTCSVAHAEST AINKTFSACALNFIPPTVKLFHSSCNPVGDT HTTIQLLCLISGYVPGDMEVIWLVDGQKATN IFPYTAPGTKEGNVTSTHSELNITQGEWVSQ KTYTCQVTYQGFTFKDEARKCSESDPRGVT SYLSPPSPL Propensity scales: The principle • The Parker hydrophilicity scale • Derived from experimental data D E N S Q G K T R P H C A Y V M I F L W 2.46 1.86 1.64 1.50 1.37 1.28 1.26 1.15 0.87 0.30 0.30 0.11 0.03 -0.78 -1.27 -1.41 -2.45 -2.78 -2.87 -3.00 Hydrophilicity Propensity scales: The principle ….LISTFVDEKRPGSDIVEDLILKDENKTTVI…. (-2.78 + -1.27 + 2.46 +1.86 + 1.26 + 0.87 + 0.3)/7 = 0.39 Prediction scores: 0.38 0.1 0.6 0.9 1.0 1.2 2.6 1.0 0.9 0.5 -0.5 Epitope Evaluation of performance • A Receiver Operator Curve (ROC) is useful for finding a good threshold and rank methods Blythe and Flower 2005 • Extensive evaluation of propensity scales for epitope prediction • Conclusion: – Basically all the classical scales perform close to random! – Other methods must be used for epitope prediction BepiPred: CBS Web server • Parker hydrophilicity scale • Hidden Markow model • Markow model based on linear epitopes extracted from the AntiJen database • Combination of the Parker prediction scores and Markow model leads to prediction score • Tested on the Pellequer dataset and epitopes in the HIV Los Alamos database Data from: J. L. Pellequer, E. Westhof, and Van M. H. Regenmortel. Correlation between the loca- tion of antigenic sites and the prediction of turns in proteins. Immunol. Lett., 36: 83–99, 1993. Ole Lund, Morten Nielsen, Claus Lundegaard, Can Kesmir and Søren Brunak. Immunological Bioinformatics. MIT press, Cambridge, Massachusetts. 2005 312 pp. ROC evaluation Evaluation on HIV Los Alamos data set Linear epitope prediction performance • Pellequer data set: – Levitt – Parker – BepiPred AROC = 0.66 AROC = 0.65 AROC = 0.68 • HIV Los Alamos data set – Levitt – Parker – BepiPred AROC = 0.57 AROC = 0.59 AROC = 0.60 BepiPred • BepiPred conclusion: – On both of the evaluation data sets, Bepipred was shown to perform better – Still the AROC value is low compared to T-cell epitope prediction tools! – Bepipred is available as a webserver: www.cbs.dtu.dk/services/BepiPred Prediction of linear epitopes Pro easily predicted computationally easily identified experimentally immunodominant epitopes in many cases do not need 3D structural information easy to produce and check binding activity experimentally Con only ~10% of epitopes can be classified as “linear” weakly immunogenic in most cases most epitope peptides do not provide antigen-neutralizing immunity in many cases represent hypervariable regions DiscoTope server • CBS server for prediction of discontinuous epitopes • Uses protein structure as input • Combines propensity scale values of amino acids in discontinuous epitopes with surface exposure • Will be available soon (not published yet) • Contact me for more information DiscoTope Andersen PH, Nielsen M, Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006 15:2558-67. Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006 2: 2. Web server output The Biological unit Prediction of conformational epitopes with DiscoTope: Refining the benchmark Characterization of the B-cell epitope area Data processing • 224 resolved 3-dimensional antigen-protein complexes – Kindly provided by Søren Padkjær • Complexes with antigens below 20 amino acids were removed = 162 • Similar interactions were removed = 109 • 2 complexes were identified as outliers and removed = 107 • 70 epitope annotated amino acids were found outside the general binding area and removed Characterization of the B-cell epitope area Spatial distribution of amino acid in epitopes • Hydrofobic center • Charged edge RSA = 0.511 Characterization of the B-cell epitope area Modeling the most likely epitope • Shape: Flat, oblong, oval shaped area • Direction: -30 to 60 degrees relative to the light to heavy chain direction • Amino acid distribution: Hydrofobic residues in the center and charged residues on the edge RSA = 0.511 Human proteome (106 peptides) on a chip Schafer-Nielsen, Søren Buus, Massimo Andretta, FP6 PepChipOmics project The parasite exports VAR2CSA to the RBC membrane which enable adhesion of parasites to CSA in the placenta IRBC Causing: Placental malaria Structural envelope of VAR2CSA with 7 domains Red blood cell membrane 1. We expressed full length 350 kD VAR2CSA and immunized rats 2. We affinity purified rat antibodies on the recombinant protein (thereby purifying IgG reacting with exposed epitopes) 3. Tested the affinity purified IgG on the PePtide array covering the entire VAR2CSA protein. Conclusion: Very few linear epitopes exposed in the VAR2CSA protein, however a number of peptides were identified. These were synthezised, coupled to KLH and used in immunization. Note 1 : The IgG before affinity purification did not reveal many more epitopes Note 2 : The same sera were tested on Pepscan array and were completely blank, ie Schafern peptide array mimicks more epitopes. Rat sera were then tested by flow cytometry to test if IgG reacts with native VAR2CSA on VAR2CSA expressing malaria parasites: MFI 18 16 14 12 10 8 MFI 6 4 2 0 pep152 KLHconjugated pep152 KLHconjugated pep153 KLHconjugated pep153 KLHconjugated Control protein Conclusion. Both peptides induce IgG that reacts with native VAR2CSA. To do: Denature VAR2CSA and induce antibodies against linear epitopes and test on array Prediction of epitopes • Cytotoxic T cell epitope: (AROC ~ 0.9) • Will a given peptide bind to a given MHC class I molecule • Helper T cell Epitope (AROC ~ 0.8) • Will a part of a peptide bind to a given MHC II molecule • B cell epitope (AROC ~ 0.7) • Will a given part of a protein bind to one of the billions of different B Cell receptors