Chapter 4 - Richsingiser.com
advertisement

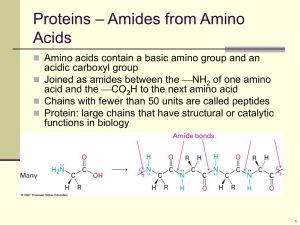
Reginald H. Garrett Charles M. Grisham www.cengage.com/chemistry/garrett Chapter 4 Amino Acids Reginald Garrett & Charles Grisham • University of Virginia Outline • What are the structures and properties of amino acids? • What are the acid-base properties of amino acids? • What reactions do amino acids undergo? • What are the optical and stereochemical properties of amino acids? • What are the spectroscopic properties of amino acids? • How are amino acid mixtures separated and analyzed? • What is the fundamental structural pattern in proteins? 4.1 What Are the Structures and Properties of Amino Acids? • Amino acids contain a central tetrahedral carbon atom • There are 20 common amino acids (you need to know all of them) • Amino acids can join via peptide bonds • Several amino acids occur only rarely in proteins • Some amino acids are not found in proteins 4.1 What Are the Structures and Properties of Amino Acids? Anatomy of an amino acid. Except for proline and its derivatives, all of the amino acids commonly found in proteins possess this type of structure. 4.1 What Are the Structures and Properties of Amino Acids? Two amino acids can react with loss of a water molecule to form a covalent bond. The bond joining the two amino acids is called a peptide bond. The 20 Common Amino Acids You should know names, structures,3-letter and 1-letter codes • • • • Non-polar amino acids Polar, uncharged amino acids Acidic amino acids Basic amino acids The 20 Common Amino Acids Some of the nonpolar (hydrophobic) amino acids. The 20 Common Amino Acids More of the nonpolar (hydrophobic) amino acids. The 20 Common Amino Acids Some of the polar, uncharged amino acids. The 20 Common Amino Acids More of the polar, uncharged amino acids. The 20 Common Amino Acids The acidic amino acids. The 20 Common Amino Acids The basic amino acids. Several Amino Acids Occur Rarely in Proteins We'll see some of these in later chapters • • • • • • Selenocysteine in many organisms Pyrrolysine in several archaeal species Hydroxylysine, hydroxyproline - collagen Carboxyglutamate - blood-clotting proteins Pyroglutamate – in bacteriorhodopsin GABA, epinephrine, histamine, serotonin act as neurotransmitters and hormones • Phosphorylated amino acids – a signaling device Several Amino Acids Occur Rarely in Proteins Several Amino Acids Occur Rarely in Proteins Several Amino Acids Occur Rarely in Proteins 4.2 What Are Acid-Base Properties of Amino Acids? • Amino Acids are Weak Polyprotic Acids • The degree of dissociation depends on the pH of the medium H2A+ + H2O ↔ HA0 + H3O+ 0 [HA ][H3O ] Ka1 [H2 A ] 4.2 What Are Acid-Base Properties of Amino Acids? The second dissociation (the amino group in the case of glycine): HA0 + H2O ↔ A− + H3O+ Ka 2 [A ][H3O ] 0 [HA ] 4.2 What Are Acid-Base Properties of Amino Acids? The ionic forms of the amino acids, shown without consideration of any ionizations on the side chain. pKa Values of the Amino Acids You should know these numbers and know what they mean • Alpha carboxyl group: pKa = 2 • Alpha amino group: pKa = 9 • These numbers are approximate, but entirely suitable for our purposes. 4.2 What Are Acid-Base Properties of Amino Acids? 4.2 What Are Acid-Base Properties of Amino Acids? Titrations of polyprotic amino acids Titration of glutamic acid A Sample Calculation What is the pH of a glutamic acid solution if the alpha carboxyl is 1/4 dissociated? [1] pH = 2 + log10 [3] • pH = 2 + (−0.477) • pH = 1.523 • Note that, when the group is ¼ dissociated, ¼ is dissociated and ¾ are not; thus the ratio in the log term is ¼ over ¾ or ⅓. Reactions of Amino Acids • Carboxyl groups form amides & esters • Amino groups form Schiff bases and amides • Edman reagent (phenylisothiocyanate) reacts with the α-amino group of an amino acid or peptide to produce a phenylthiohydantoin (PTH) derivative. • Side chains show unique reactivities • Cys residues can form disulfides and can be easily alkylated • Few reactions are specific to a single kind of side chain Reactions of Amino Acids Cysteine residues react with each other to form disulfides. This reaction is an oxidation-reduction reaction. Green Fluorescent Protein A jellyfish (Aequorea victoria) native to the northwest Pacific Ocean contains a green fluorescent protein. GFP is a naturally fluorescent protein. Genetic engineering techniques can be used to “tag” virtually any protein, structure, or organelle in a cell with GFP. The luminescent chromophore of GFP lies in the center of a βbarrel protein structure. Green Fluorescent Protein The prosthetic group of GFP is an oxidative product of the sequence –FSYGVQ-. Stereochemistry of Amino Acids • All but glycine are chiral • L-amino acids predominate in nature • D,L-nomenclature is based on D- and Lglyceraldehyde • R,S-nomenclature system is superior, since amino acids like isoleucine and threonine (with two chiral centers) can be named unambiguously Stereochemistry of Amino Acids Spectroscopic Properties • All amino acids absorb at infrared wavelengths • Only Phe, Tyr, and Trp absorb UV • Absorbance at 280 nm is a good diagnostic test for amino acids • NMR spectra are characteristic of each residue in a protein, and high resolution NMR measurements can be used to elucidate three-dimensional structures of proteins Spectroscopic Properties Figure 4.10 The UV spectra of the aromatic amino acids at pH 6. Spectroscopic Properties Proton NMR spectra of several amino acids. 4.7 What is the Fundamental Structural Pattern in Proteins? • Proteins are unbranched polymers of amino acids • Amino acids join head-to-tail through formation of covalent peptide bonds • Peptide bond formation results in release of water • The peptide backbone of a protein consists of the repeated sequence –N-Cα-Co• “N” is the amide nitrogen of the amino acid • “Cα” is the alpha-C of the amino acid • “Co” is the carbonyl carbon of the amino acid 4.7 What is the Fundamental Structural Pattern in Proteins? Peptide formation is the creation of an amide bond between the carboxyl group of one amino acid and the amino group of another amino acid. The Peptide Bond • • • • is usually found in the trans conformation has partial (40%) double bond character N partially positive; O partially negative has a length of about 0.133 nm - shorter than a typical single bond but longer than a double bond • Due to the double-bond character of the peptide bond, the six atoms of the peptide bond group define a plane – the amide plane The Peptide Bond The trans conformation of the peptide bond. 4.7 What is the Fundamental Structural Pattern in Proteins? The peptide bond has partial double bond character. One of the postulated resonance forms is shown here. 4.7 What is the Fundamental Structural Pattern in Proteins? Figure 4.16 (b) The peptide bond has partial double bond character. One of the postulated resonance forms is shown here. 4.7 What is the Fundamental Structural Pattern in Proteins? Figure 4.16 (c) The peptide bond is best described as a resonance hybrid of the forms shown on the two previous slides. 4.7 What is the Fundamental Structural Pattern in Proteins? The coplanar relationship of the atoms in the amide group is highlighted here by an imaginary shaded plane lying between adjacent α-carbons. “Peptides” • • • • • • Short polymers of amino acids Each unit is called a residue 2 residues - dipeptide 3 residues - tripeptide 12-20 residues - oligopeptide many - polypeptide “Protein” One or more polypeptide chains • • • • • • One polypeptide chain - a monomeric protein More than one - multimeric protein Homomultimer - one kind of chain Heteromultimer - two or more different chains Hemoglobin, for example, is a heterotetramer It has two alpha chains and two beta chains Proteins - Large and Small • Insulin - A chain of 21 residues, B chain of 30 residues -total mol. wt. of 5,733 • Glutamine synthetase - 12 subunits of 468 residues each - total mol. wt. of 600,000 • aConnectin (a muscle protein) - MW 2.8 million • bConnectin - MW of 2.1 million, with a length of 1000 nm - it can stretch to 3000 nm Proteins - Large and Small Proteins - Large and Small From Table 4.2 Size of protein molecules. Molecular weights: Insulin, 5,733 Cytochrome c, 12,500 Ribonuclease, 12,640 Lysozyme, 13,930 Myoglobin, 16,980 Proteins - Large and Small From: Table 4.2 Size of Protein Molecules Molecular weights: Hemoglobin, 64,500; Immunoglobulin, 149,900; Glutamine synthetase, 600,000. The Sequence of Amino Acids in a Protein • • • • Is a unique characteristic of every protein Is encoded by the nucleotide sequence of DNA Is thus a form of genetic information Is read from the amino terminus to the carboxyl terminus • Questions: 1-9, 13-16, 18