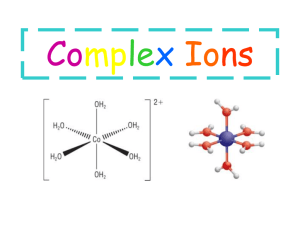
metal ions
advertisement

Biocomplexes Metal binding ability of biomolecules Donor atoms in biological systems hetero atoms with electron pairs: O, N, S, (Se) - oxygen donor ligands: alcohols: R-OH, (e.g. carbohydrates) ethers: R-O-R (e.g. carbohydrates) carbonyl compounds: -CO- (e.g. proteins) phenols: Ar-OH (e.g. polyphenols) carboxyilic comounds: -COOH (e.g. carboxylic acids, amino acids) O-heterocycles: (furane, ..) - nitrogen donor ligands: amines: R-NH2 (e.g. amino acids) amides: -CONH- (e.g. peptide bond) N-heterocycles: (pyrrole, pyridine, imidazole,...) - sulphur donor ligands: thiol compounds/disulphides: RSH/R-S-S-R (e.g. cysteine-cystine) thioethers: R-S-R (pl. methionine) sulphur containing heterocycles: (pl. tiophene,..) Main groups of bioligands and their complex forming properties - Coordination chemistry of amino acids: O H2N CH C OH a/ amino acids without other functional group: Gly, Ala, Phe,.... 5-membered chelate ring (NH2,COO-)-coordination CH3 O H2 N CH C CH2 O OH H2 N CH C CH2 SH N NH His Cys OH b/ amino acids with 3 functional groups: formation of 2 chelate rings alcoholic-OH: Ser, Thr carboxylate: Asp, Glu, amide: Asn, Gln, amine: Lys, Orn, thio: Cys, Met, imidazole-N: His - Coordination chemistry of peptides R1 CH H 2N C O NH CH C R2 O NH CH C R3 O NH CH COOH R4 1. ML, ML2,..................MLn (NH2, CO)- or COO— coordination, the latter is less favoured 2. ML MLH-1 (NH2,CO) (NH2,N-,CO) MLH-2 (NH2,N-,N-,CO) MLH-3 (NH2,N-,N-,N-) Metal ion promoted amide deprotonation and coordination Cu(II), Ni(II), Pd(II), ……... Formation of joint chelate systems is preferred O R2 CH C O C CH R1 N- N- M2+ NH2 N- R3 CH CH MH-3L R4 C O COO- 1. Coordination of the Peptide skeletone in 4N complexes (joint, five membered chelates) 2. Site specific role of R1, R2,.. Side chains (His, Cys) - coordination chemistry of proteins: (the ligands have predetermined structure) Chelate formation has only secondary importance Independent coordination of the side-chain donor atoms e.g. structure of carboanhydrase: His 96 H 2O Zn2+ His 119 His 94 Carboanhydrase Thr 199 His 64 Glu 106 His 96 H2O Zn2+ His 119 His 94 Gln 92 Glu 117 Carboanhydrase - coordination chemistry of proteins: the proteins significantly change their structure due to metal ion coordination e.g. development of the zinc-finger structure: - Coordination chemistry of nucleic acids and their building blocks NH2 AMP N N N O - O P O H N H H H OH purine (adenine/guanine) pirimidine (cytosine/uracil) - sugar: ribose or deoxy-ribose O O- Potential binding sites: - nucleic bases: - phosphate H Characteristics: no chelate formation → macrochelate or loop formation is possible Metal ion selectivity: Nucleo bases: „soft” metal ions: e.g. Pt biological role Phosphate: „hard” metal ions: e.g. Ca, Mg, Al,... Difference in DNA/: cis-OH groups → (chelate formation) - Metalloporphyrins and related compounds Composition/Structure: N-donor containing, natural macrocycles complex forming: porphine, chlorin-, corrin-, corphin-ring containing compounds 4 pyrrole (or indole) rings in joint conjugated or partly conjugated system (most important: tetrapyrrole compounds) Stability of their metal complexes: Differs from the usual stabilty sequences Ratio of the inner hole and the size of the metal ion has primary importance. Metallation is a catalytic process. Metalloporfirinek - Metalloporphyrins M2+ + H2P N N M MP + 2H+ N N Stability Stabilitásitrend sor: Mg(II) < Zn(II) < Cu(II) < Fe(II) < Ni(II) < Pd(II) < Pt(II) All coordination siteshely are saturated N = 4 (Ni(II), Pt(II), Pd(II)) - minden koordinációs foglalt Axial binding sites N= 6 (Fe(II), Co(II), Mg(II), Zn(II) - axiális kötõhely Complexation with other bioligands - carbohydrates: numerous –OH groups in acidic/neutral milieau weak metal ion binders might be suitable for administration of metal ions (medicinal application) -derivatives: aldolic acids amino sugars, - lipids, oils: contain few functional groups very weak complexation Summary: Strongest metal ion binders: proteins and macrocyclic ligands → enzymes, coenzymes, prosthetic groups Further interactions: redox and acid-base interactions with vitamines, hormones and many other biomolecules Ellenőrző kérdések 1. 2. 3. 4. 5. 6. Mi az alapvető különbség az aminosavak és az oligopeptidek fémionkötő képessége között? Mi a jellemző a fehérjék fémion koordinációjára? Mely oldallánc donorcsoportok játszanak meghatározó szerepet a fémionok megkötésében a fémtartalmú fehérjékben? Adjon példákat! Jellemezze a nukleinsavak és nukleotidok fémion koordinációját! Miért gyenge a szénhidrátok fémion megkötő képessége? Hogyan fokozható ez? Milyen biomolekulákban fordul elő a tetrapirrol váz? Röviden jellemezze ezeket! Basic coordination chemistry 1. Basic terms: (see advanced inorganic chemistry) formation: Lewis acid-base reaction ligand: base metal ion: acid equilibrium:stepwise (successive) kinetics: labile and inert complexes coordination number: 2 – (10) with essential elements: (2), 4, 6 coordination geometry: 4 – tetrahedral/square planar, 6 – octahedral effects of complex formation: colour and magnetic feature (crystal field theory) changes in the redox potential: (Fe3+/Fe2+ and Cu2+/Cu+ systems) 2. Complex formation in biological systems (multi)component systems pM + sH + qA + rB MpAqBrHs Types of complexes: biner (parent): MA, ........... MAn polynuclear: MnA (A – bridging ligand) protonated: MHA (multifunctional ligand) MH-1A (coordinated H2O – hydroxocomplex or „A”-ligand metal ion induced deprotonation terner (mixed ligand/metal complexes): MAB Reactions of complexes: acid-base: liberation/uptake of protons redox: redox reactions of the metal ion or coordinated ligands other reactions of coordinated ligands: templates, changes in conformation, etc. Hard-soft classification of the biologically important metal ions and ligands The preferential coordination of metal ions to bioligands/biodonors generally can be well explained by the hard-soft acid-base theory. The basic features of hard and soft acids and bases Acids Hard Low Low Higher Small Ionic bond Soft Polarizability Electronegativety Positive charge Size Chemical interactions High High Lower Large Covalent or p-bound Bases Hard Low High Higher Small Ionic bond Soft Polarizability Electronegativety Negative charge Size Chemical interactions High Low Lower Large Covalent or p-bound The general rule is that hard acids makes strong interactions with hard bases and soft acids with soft bases. Accordingly, hard acids, like Ca2+, Fe3+, Al3+ prefer the oxygen-, fluorine- and partly N donor atoms, while the soft acids, like Cu+, Pt2+, Hg2+ and Cd2+ prefer the sulphur, phosphorous, and iodine donors in forming coordination compounds. Splitting of d-orbitals in different fields Because of the different „shape” of the d orbitals the energy degeneracy of the orbitals is lifted. Orbitals in a given geometry directed towards the ligands (electron pairs) will have higher energy and will be able to form bonds, while those not directed to ligand lone electron pairs will occupy lower energy and will be able to form p bonds. Distribution of electrons on the d-orbitals When the electrons will be redistributed among the d orbitals the sequence will be determined by the relation between the crystal field splitting energy () and the spin pairing energy (P). The figure shows the possible electron configurations in the case of octahedral geometry. Common geometries for 2-6 coordinate metal ions Cation Na+ K+ Mg2+ Ca2+ Mn2+ (d5) Coord number 6 6-8 6 6-8 6 Geometry Octahedral Flexible Octahedral Flexible Octahedral 6 4 6 Tetrahedral Tetrahedral Octahedral 4 6 Tetrahedral Octahedral 4 Square planar 6 Octahedral 4 Square planar Ni2+ (d8) 6 4 Octahedral Tetrahedral Cu1+ (d10) 4 Tetrahedral Mn3+ (d4) Fe2+ (d6) Fe3+ (d5) Ca2+ (d7) Suqare planar Cu2+ (d9) Zn2+ (d10) 6 Octahedral 4 Tetrahedral 5 Square pyramidal Biologic ligands O, ether, hydroxyl, carboxylate O, ether, hydroxyl, carboxylate O, carboxylate, phosphate O, carboxylate, carbonyl (phosphate) O, carboxylate, phosphate N, imidazole N O, carboxylate, phosphate, hydroxyde S, thiolate O, carboxylate, alkoxide, oxide, phenolate, N, imidazole N, porphyrin S, thiolate O, carboxylate, alkoxide, oxide, phenolate, N, imidazole N, porphyrin S, thiolate N, imidazole N O, carboxylate N, imidazole N S, thiolate N, imidazole N, polypirrol (F-430) Rare S, thiolate, thioether N, imidazole N S, thiolate, thioether N, imidazole N O, carboxylate N, imidazole N O, carboxylate N, imidazole N O, carboxylate, carbonyl S, thiolate N, imidazole N O, carboxylate, carbonyl N, imidazole N Standard redox potential of several iron complexes The following basic conclusions can be drawn from the data in the above Table: (i) a decrease in the redox potential means stabilisation of the FeIII state as compared to FeII. That is, in the presence of hydroxide, cyanide, or oxalate ions FeII can be oxidised to FeIII or FeIII can hardly be reduced to FeII. In basic solution weak oxidising agents, like molecular oxygen, can oxidise FeII to FeIII. (ii) an increase in the redox potential means that the FeII state is stabilised as compared to the FeIII. In the presence of 2,2’-dipyridyl or 1,10-phenantrolin FeII can be oxidised to FeIIIonly by very strong oxidising agents. Redox potential of several copper complex Redox system Cu(alanine)22+ + e- → Cu(alanine)2+ Cu(glycine)22+ + e- → Cu(glycine)2+ Cu2+ + e- → Cu+ CuL22+ + e- → CuL2+ Cu(pyridine)22+ + e- → Cu(pyridine)2+ Cu(imidazole)22+ + e- → Cu(imidazole)2+ Cu(CN)2 + e- → Cu(CN)- E0(V) -0.130 -0.160 +0.167 +0.243 +0.270 +0.345 +1.103 (L = 2-methyl-thioethyl-amine) In aqueous solution standard redox potential of the Cu2+ + e- Cu+ system is +,167 V. This suggests that in the absence of any complexing agents the Cu2+ ions are more stable. Depending on the type of the ligands either the CuII, or the CuI state can stabilised. From the data in the above Table it can be concluded that the lower oxidation state of copper the CuI can be stabilised in the presence of aromatic N-compounds (pyridin, imidazol), furthermore sulphur containing compounds, while the CuII state can be stabilised by N or O containing ligands (e.g. amino acids, such as alanin). Exchange rates for inner sphere water molecules In general, it can be said that the exchange rate is higher for the less highly charged less strongly bound metal ions, than the more highly charged metal ions. The highly inert first row Cr3+ és Co3+ metal ions practically have no biological importance. Similarly, the second and third row transition metal ions have less biological importance too. pK of various ligands in the absence and presence of biologically relevant metal ions Ligand and reaction Metal ion lg K Methods in bioinorganic chemistry I. 1. Determination of the composition: - solid phases: elemental analysis - solution equilibrium studies (determination of the stability constants) (potentiometry, other techniques → measurements of some of the free components) - mass spectrometry (ESI-MS) 2. Kinetics: - slow substitution reactions (inert complexes): classical analytical methods → measurements of the concentration of some of the components) - fast substitution reactions (labile complexes): stopped flow, T- jump and realxation methods (e.g. NMR). Physical methods in bioinorganic chemistry II. 3. Structural investigating methods: a/ optical spectroscopic methods: - UV-Vis spectrophotometry (d-d transitions, charge transfer (CT) and ligand transitions) - circular dicroism (CD, optical active compounds) b/ magnetic methods: - magnetic momentum (low and high spin complexes) - ESR (EPR) spestroscopy (e.g. Cu2+, Mn2+, VO2+,...) - NMR spectroscopy (1H, 13C, ligand peaks) multinuclear: 15N, 17O, 19F, 27Al, 51V, 113Cd, 195Pt,.. c/ other methods: - Mössbauer spectroscopy: (pl. Sn, Fe...) - X-ray diffraction - mass spectrometry Enzymes I. 1. Definition: Catalysts of biological systems 2. Importance: chemical process: A B the catalyst accelerates the time reaching the equilibrium state, but equilibrium concentrations [A], [B] do not change. Biological system: stationer equilibrium (steady state) → A B C D ............... → E1 E2 E3 [A]stat, [B]stat, [C]stat = f(E1,E2,…En) The concentrations measured at the stationer equilibrium state are not the same as those in the thermodynamic equilibrium, but depend upon the reaction rates (enzymes). → The system is in continous change, it „goes” to the equilibrium state. Enzymes II. 3. Naming enzymes: process + ase e.g. peptidase (hydrolysis of peptides) carboxypeptidase (from the direction of the C-terminus) ( enzyme catalogue: EC x.y.z.w. e.g. EC 6.3.1.2. glutaminsynthetase) 4. Classification of Enzymes: Class oxidoreductases transferases hydrolases liasese ligases isomerases type pf reaction redox reactions transfer of atom or group of atoms hydrolysis non hydrolytic cleavege linking groups together isomeric trasformation Enzymes III. 5. Selectivity: usually high function specificity: catalysis of a given reaction type (e.g. carboxypeptidase) substrate specificity: catalysis of a given range of substrates within the function specificity (e.g. carboxypeptidase A: only with substrates with hydrophobic side chain) 6. Composition: simple or complex proteins simple protein: M ≥ 10.000 ( ≥ 100 amino acid) complex protein: protein + prosthetic group or coenzyme prosthetic group: reversible non separable (e.g. hem, biotin, metal ions, e.g. Cu, Fe,..) coenzyme: existing biomolecule (e.g. NAD, ATP, Mo-co, metal ions, e.g. Ca, Mg, Zn,...) [ribozymes: RNA based enzymes] Enzymes IV. 7. Metalloenzymes: ~ 30 % of the enzymes - Binding of the metal ions: prosthetic group (e.g. hem, Fe, Cu,...) coenzyme: (e.g. B12, Mo-co, Ca, Mg, Zn, .....) - Role of the metal ions: active centre: direct interactions with the substrate. characteristics: distorted and unsaturated coordination geometry (high energy state) (e.g. carbonanhydrase (Zn), carboxypeptidase(Zn), superoxide dismutase(Cu)) structure maker (stabilizer): the metal ion fixes the conformation of the protein, usually saturated coordination geometry. (e.g. alcohol dehydrogenase(Zn), superoxide dismutase(Zn) Kinetics of enzyme reactions I. 1. Kinetic model: Michaelis-Menten Thesis: The initial rate (vo) of the enzyme catalysed reactions show saturation curve in the function of the concentration of the substrate ([S]). vo Vmax Vmax/2 KM [S] Kinetics of enzyme reactions II. The general equation describing the previous function: a S vo b S E+S k1 a = Vmax b = KM To explain this the stationary equilibrium (steady state) has to be applied: ES k-1 k2 → E+P i.e. the product (P) is formed through an enzyme-substrate complex (ES). If the concentration of the substrate is sufficiently high ([S] >> [Eo]) Concentration of ES is constant in time, i.e. the rate of its formation and decomposition equals: dES k1 E S dt dES (k 1 k 2 ) ES dt Kinetics of enzyme reactions III. Assuming the stationary equilibrium: dES dES dt dt k1 E S (k 1 k 2 ) ES Taking into account the equations expressing of total enzyme concentrations and the rate equation of product formation: v o k 2 ES E Eo ES By substituting and rearranging the above equations: k 2 Eo S vo k 1 k 2 S k1 The initial rate is at maximum, when [ES] = [Eo] Vmax = k2·[Eo], i.e. in the rate equation: a k 2 Eo Vmax and b = KM k 1 k 2 k1 Inhibition of enzyme reactions I. 1. Reversible inhibition: (I: inhibitor, E: enzyme, S: substrate) a/ competitiv inhibition: the inhibitor and the substrate compete for the active centres of the enzyme I→E but I → ES KM increases, but Vmax ≠ f(I) b/ non-competitiv inhibition: the inhibitor interacts also with the enzyme-substrate complex (a Vmax also decreases) Inhibition of enzyme reactions II. 2. Irreversible inhibition: Strong (covalent) interactions between the inhibitor and the Enzyme, which can not be cleaved by the substrate. If [I] > [E] activity of the enzyme can be completely stopped. Heavy metal ions and chelators can be frequent inhibitors. Mechanism of enzyme reactions (metalloenzymes) Mechanism of the metalloenzyme catalysed reactions can be grouped in three main types: (L: substrate, activator or inhibitor) 1. Ligand bridged (or substrat bridged) There is no direct interaction between the metal ion and the enzyme, But it is necessary for the activation of the enzyme-substrate complex. 2. Metal bridged a/ The substrate is in direct interaction only with the metal ion (occurs very rarely) b/ The substrate is in interactions with the enzyme and the metal ion too (mpost frequent case) 3. Enzyme bridged There is no direct interaction between the metal ion and the substrate, But binding of the metal ion to the protein is necessary to the catalytic activity (structure maker) (e.g. alcohol dehydrogenase)