Nitrogen Metabolism
advertisement

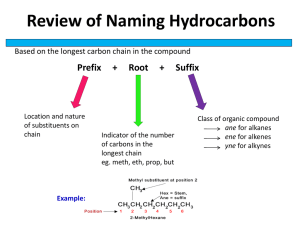
Nitrogen Metabolism • • • • • • Protein degradation and turnover Amino acid degradation and urea cycle Nitrogen cycle Nitrogen fixation Amino acid biosynthesis Amino acid derivatives How Much Protein? • A 70 kg person (154 lb) typically consumes 100 g protein per day • To stay in nitrogen balance that person must excrete 100 g of N products per day • The body makes 400 g of protein per day and 400 g are broken down • 300 g of amino acids recycled into new protein, 100 g are degraded • Total protein = 500 g/day, 400 g degraded, 400 resynthesized and 100 g catabolized Characteristic of Proteins in Cells • Synthesized and degraded constantly Turnover • Turnover may be minutes, weeks or longer • Synthesis requires essential and non essential amino acids • Degradation is programmed and regulated • Control point enzymes most labile; constitutive most stable • Nutritional state and hormones affect degradation rates (glucocorticoids, insulin, etc.) The half-life of proteins is determined by rates of synthesis and degradation dC Rate of Turnover = dt = KS - KDC A given protein is synthesized at a constant rate KS A constant fraction of active molecules are destroyed per unit time KS is the rate constant for protein synthesis; will vary depending on the particular protein C is the amount of Protein at any time KD is the first order rate constant of enzyme degradation, i.e., the fraction destroyed per unit time, also depends on the particular protein Steady-state is achieved when the amount of protein synthesized per unit time equals the amount being destroyed dC = 0 KDC = KS dt t 1/2 = 0.693 KD C Protein concentration (enzyme activity) Stop protein synthesis, measure rate of decay Hours after stopping synthesis Steps in Protein Degradation Transformation to a degradable form (Metal oxidized, Ubiquination, N-terminal residues, PEST sequences) Lysosomal Digestion ATP 26S Proteasome digestion AMP + PPi Proteolysis to peptides KFERQ 7 type, 7 type subunits 8 residue fragments Ubiquination N-end rule: PEST: DRLKF: 2-3 min AGMSV: > 20 hr Rapid degradation Glycine at C terminal of Ubiquitin Ubiquitin COOATP Ubiquitin activating enzyme HS AMP + PPi E1 O C S Activation of Ubiquitin Ubiquitin conjugating enzyme 20 or more per cell NH3+ E1 HS E2 3 HS E1 C S Ubiquitin ligase E2 O C N O C Poly Ubiquitin Ubiquination Page 1075 SH NH3+ H3N+ O 3 E2 ATP NH AMP + PPi Degraded protein E3 O N C O N C Ubiquitinspecific proteases (26S proteasome) + Ubiquitin Cervical Cancer Human Papilloma virus (HPV) Activates the E3 that catalyzes ubiquination of p53 tumor suppressor and DNA repair enzymes (occurs in 90% of cervical cancers) Mutated DNA is unchecked and allowed to replicate P472 19S 20S Catalysis in beta 19S Subunits 26S Proteasome (2000 kD) 7 alpha 7 beta Opening for ubiquinated protein to enter 8-residue peptides diffuse out Amino Acids Amine Group Glutamate Urea Carbon Skeleton Biosynthesis Amino Acids Amino Acid Derivatives Degradation CO2 + H2O -Ketoglutarate-Glutamate COOC=O CH2 CH2 COO-Kg Amine group acceptor COO+ H3N-C-H CH2 Amine group donor CH2 COOL-glutamate AA1 + -KG -ketoacid + glutamate acceptor donor Amino transferases Requires pyridoxal-5’-phosphate CH2OH Vitamin B6 HO H 3C CH2OH Pyridoxine N O Cofactor (N acceptor) C HO H3 C H CH2OP N CH2NH2 Cofactor (N donor) HO H3 C Pyridoxal-5’-PO4 CH2OP N Pyridoxamine-PO4 Alanine-Pyruvate Aminotransferase COO+ H3N-C-H + CH3 O C HO H3 C COOC=O CH2 CH2 COOforward reverse H HO H3C CH2OP N O C CH2NH2 CH2OP N COO+ H3N-C-H C=O + CH2 CH3 CH2 COO- COO- HO H3 C H CH2OP N Mechanism Alanine In Glutamate Pyruvate -Ketoglutarate Out Enz-CHO (E-B6-al) Enz-NH2 (E-B6-am) In Enz-NH2 Out Enz-CHO Ordered Ping-Pong Mechanism Glutamate Metabolism COO+ H3N-C-H CH2 + NAD(P)+ + H2O CH2 Glutamate COOdehydrogenase COOC=O CH2 + NAD(P)H + H+ + NH4+ CH2 COO- Urea cycle Forward Reaction Reverse Reaction specific for glutamate specific for -ketoglutarate requires NAD+ requires NADPH delivers NH4+ to urea cycle Fixes NH4+, prevents toxicity Glutamine Metabolism COOCOO+ + H3N-C-H H3N-C-H + ATP + NH4+ CH2 + ADP + Pi CH2 Glutamine CH2 CH2 L-glutamine Synthetase C=O COONH2 H2O COOGlutaminase + H3N-C-H COO CH2 + H3N-C-H CH2 + NH4+ Glutamate-PO4 CH2 C=O intermediate CH2 = OPO3 Urea COO- Overall Scheme Using Alanine as an Example Amino transferase with pyridoxal-5’-PO4 Alanine Pyruvate -ketoglutarate glutamate Glutamate dehydrogenase with NAD+ NH4+ Glutaminase with H2O glutamine Urea Glutamate and glutamine are the only donors of NH3 to the Urea Cycle The Urea Cycle 1. Occurs in the liver mitochondria and cytosol 2. Starts with carbamoyl-PO4 3. Ends with arginine 4. Requires aspartate 5. Requires 3 ATPs to make one urea Synthesis of Carbamoyl-PO4 NH4+ + HCO3- + 2 ATP O H2N O C ~ O-P-O + 2 ADP + Pi O High energy bond Carbamoyl phosphate Synthetase I Citrulline Carbamoyl-PO4 Urea Cycle Ornithine + NH3 CH2 CH2 CH2 HC COOH3N Arginine H2O O C H2 N NH2 Urea Aspartate ATP Argininosuccinate NH2 + H2N=C NH CH2 CH2 CH2 HC COOH3N Reactions of Urea Cycle COOH3N+-C-H O CH2 CH2 CH2 + NH 3 COOH3N+-C-H + H2N C CH2 CH2 CH2 NH OPO3 Carbamoyl-PO4 O=C Ornithine CH2 CH2 CH2 NH O=C NH2 H3N+-C-H COO+ Citruline COO- COOH3 + OPO3= NH2 Mitochondria N+-C-H Cytosol ATP CH2 + H-C-NH3 COO- L-Aspartate ADP + Pi CH2 CH2 COO- CH 2 CH2 NH H-C-N =C COO- NH2 Argininosuccinate Cytosol COOH3N+-C-H COOCH2 CH2 CH2 COO- CH2 CH2 NH CH2 CH2 NH CH H2N+ =C H-C-N =C COO- COOH3N+-C-H NH2 + HC COOFumarate NH2 L-Arginine COO- COO- COO- CH2 CH2 CH2 H-C-NH3 C=O H C-OH COO- COO- + L-Aspartate Oxaloacetate COOL-Malate COOH3N+-C-H COOH3N+-C-H CH2 H2O CH2 CH2 CH2 NH CH2 CH2 + NH3 NH2 Ornithine H2N+ =C O + C H2N NH2 Urea L-Arginine Return to Mitochondria