The Main MOE Windows:
advertisement
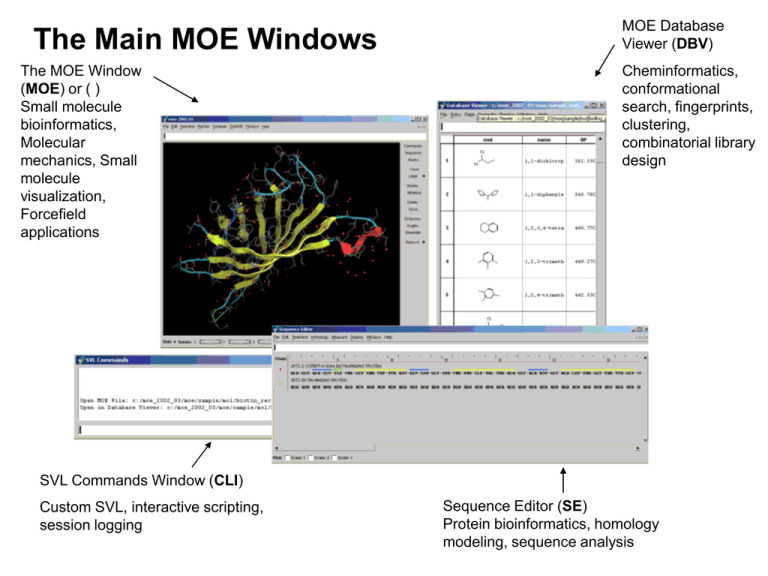
The Main MOE Windows The MOE Window (MOE) or ( ) Small molecule bioinformatics, Molecular mechanics, Small molecule visualization, Forcefield applications MOE Database Viewer (DBV) Cheminformatics, conformational search, fingerprints, clustering, combinatorial library design SVL Commands Window (CLI) Custom SVL, interactive scripting, session logging Sequence Editor (SE) Protein bioinformatics, homology modeling, sequence analysis Intro 1: The MOE Window Used for: • Building small molecules • Molecular mechanics • Structure-based drug design • Docking • SCF Calculations • Molecular dynamics • Flexible alignment • PH4 elucidation • Conformational searching Intro 2: The Sequence Editor Used for: • Protein bioinformatics • Sequence alignment • Homology searching • Homology modeling • Target family analysis • RCSB download • Consensus modeling • PDB searching Intro 3: The MOE Database Viewer Used for: • Cheminformatics • QSAR • Conformation search output • Dynamics output • Flexible alignment output • Docking output • Clustering • Fingerprints • Similarity search • Diverse subset • Data correlation • Combinatorial library design • R-group preparation • PH4 searching • Washing / Preprocessing Intro 4: The SVL Commands Window Used for: • Custom SVL • Interactive scripting • Session logging Layout of Course 1. Main MOE Window: a) Opening/saving files b) Building molecules c) Rendering 2. Introduction to the Sequence Editor 3. Introduction to the Database Viewer 4. a) Basics of molecular mechanics and conformational searching b) Basics of cheminformatic analysis with the Database Viewer Comments on SVL 1. Structure of the MOE Window Task Cancel Button Main Menu Commands RHS Button Bar SVL Command Line Popup menu 3D Rendering Area Footer Pager Bar MOE Menu Command Conventions • Commands from the MOE Window are preceded by MOE or () (Render | Backbone | Color | Chain Color) or (MOE | Render | Backbone | Color | Chain Color) Mouse Conventions in MOE General Mouse Actions LEFT 3-Button Mouse – 2-Button Mouse Mapping - Selecting objects, menu commands MIDDLE - Rotating, translating moving objects RIGHT - SE and DBV Popup menus Press and release <Alt> key Input / Output File Formats in MOE • MOE can read various input formats, e.g. MOE, PDB, SD etc. • A variety of export file formats are possible, e.g. MOE, Tripos MOL2 etc. • Picture files may be generated for publications or presentations. Opening Files in MOE – (File | Open) Path Text Field Current Path/ Directory Change Working Directory (CWD) Enforce File Type ‘..’ go up a Directory Recent Directories List Directory/ File List Open file into MOE Open file in text editor Operations to Perform on selected file(s) Exercise: Opening a File 1. Open the File Open panel (File | Open). 2. Use the pull-down menu to switch to the $MOE/sample/mol directory. 3. Find the file $MOE/sample/mol/sulph_quin.moe 4. Open MOE file into the MOE Window by either: a) Selecting the file and clicking OK or Open in MOE. b) Double left-clicking on the filename. Exercise: Opening a File (cont.) 5. Center the View (Render | View or RHS | View). 6. Render the molecule in stick mode (Render | Stick). Exercise: Manipulate molecule in 3D Window Left Click: Select atoms one at a time Middle click: Change center of rotation Right Click: Popup menu Left Click in Empty Space: To de-select to clear Middle Drag: Left Drag: Selection Box XY Rotate Ctrl Middle Drag or Scroll wheel Zoom in/out Exercise: Render molecule • Render as ball and stick, label all atoms by name, and show H-Bonds 1. MOE | Render | Draw | Hydrogen Bonds 2. Mode | Ball and Stick 3. Label | Name 4. Label | Clear Exercise: Saving a picture (1) 1. Choose MOE | File | Save 2. Click on MkDir to create a new directory called ‘course’ 3. Click on Set CWD to set as the new working directory Exercise: Saving a picture (2) 4. 5. 6. 7. Choose Save ‘Picture’ Enter the filename ‘sulph_quin.png’ Choose Format ‘PNG’ Finally click on Save Rendering 2D Depicted Molecules 1. MOE | Edit | Automatic | Depict as 2D 2. Press ‘Export Bitmap’ to save picture as ‘sulph_quin2.png’ 3. Press OK 4. Press Close Drawing Molecular Surfaces • Create Molecular Surfaces via the Molecular Surface panel (MOE | Compute | Surfaces and Maps) • Manage surfaces and other graphics objects with the Graphic Object Manager (MOE | Window | Graphic Objects) Molecular Surfaces and Maps • • • • Tool for active site analysis – Integration of three applications: Molecular Surfaces, Contact Preference Maps and new Electrostatic Maps – Easy control of definition for atom sets – Automatic handling of surface names for easy comparisons Build molecular surfaces – Gaussian, Connolly and VDW – Color by various properties Predict contact preferences – Plot knowledge based potentials for hydrophilic and hydrophobic contacts Calculate electrostatic maps – Plot electrostatically preferred positive, negative and neutral regions Exercise: Drawing Molecular Surfaces (1) 1. Draw a surface around the inhibitor by first choosing (MOE | Compute | Surfaces and Maps) using the default options 2. Press Apply Exercise: Outputting the system… to the printer To print the current MOE 3D window choose MOE | File | Print... Printer or Postscript file Landscape/Portrait Header Footer Exercise: Outputting the system… to various formats 1. Save the current MOE 3D window. MOE | File | Save... 2. Enter filename to save ‘my_sulph_quin.moe’ 3. Choose Save: Molecule and Format: .moe. To save surface, select Graphics:All 4. Click on Save Exercise: Close System and Open Builder 1. Close the current system (MOE | RHS | Close) 2. Open the Builder (MOE | RHS | Builder) The Molecule Builder Other atom types, including dummy atom at centroid Edit or Add Element Edit Ionization State Fragment substitution buttons Edit Chirality Enter Fragment SMILES string Edit: 1. Compound Name 2. Bond Length 3. Bond Angle 4. Torsion Undo button Library of functional groups Exercise: Build a molecule Press Select C Select C Press Press Exercise: Build a molecule (cont.) Press Shift-select 2 C <Shift> Shift-select 2 C Press <Shift> Exercise: Build a molecule (cont.) Select H Press Select H Press 4 times Exercise: Build a molecule (cont.) Press Press Select H Press Deselect H Exercise: Energy Minimize RHS | Minimize Save Molecule 1. Save the current MOE 3D window as a MOE file. MOE | File | Save... Enter filename to save ‘my_first_molecule.moe’ Save ‘Molecule’ Choose Format ‘MOE’ 2. Press Save Protein and Carbohydrate Builders MOE | Edit | Build | Protein or SE | Edit | Protein Builder MOE | Edit | Build | Carbohydrate Selecting Atoms with the Left Mouse Button <Ctrl>-Left Click: Auto-extend selection to residue Left Click: Select atoms one at a time <Ctrl> <Shift> <Shift>-Left Click: Add to / toggle atom selection Left Drag: Selection Box Note on <Ctrl>-Left Click: There is only one residue in the built molecule, so the whole molecule will be selected. This will be revisited later. Exercise: Selecting Atoms with the Left Mouse Button 1. Use Left-Click to select atoms one at a time. 2. Use Left-Drag to draw a selection box. 3. Use <Shift>-Left Click extend/toggle atom selections. 4. Use <Ctrl>-Left Click to select entire residues. 5. Left-mouse click in empty space to de-select any atoms. Exercise: Fixing/Unfixing Atoms (Edit | Potential | Fix / Unfix) Fixing atoms: 1. Left mouse drag to select the atoms to be fixed. 2. Fix the atoms with the command (Edit | Potential | Fix) . Fixed Atoms do not move until unfixed. Unfixing atoms: 1. Select the atoms to be unfixed. Use (Selection | Potential | Fix) to select all the fixed atoms. 2. Unfix the atoms with the command (Edit | Potential | Unfix). Once unfixed the atoms may move. Exercise: Fixed atoms and rotatable bonds If two atoms in a rotatable bond are selected <Alt>-Left Drag will rotate about the bond. If no atoms are fixed, the small group rotates by default. <Alt> Drag The larger group can be forced to rotate by fixing an atom in the smaller group FIX this atom <Alt> Drag Meters and Measurement (Edit | Measure) or (RHS | Measure…) • Choose Distances to measure and display the distance between two atoms. • Choose Angles to measure and display the angle between three atoms. • Choose Dihedrals to measure and display the dihedral angle between four atoms. Meters – Creating and Removing To create a meter, choose MOE | Edit | Measure | Distances. To remove it, select the atoms involved, and use RHS | Remove | Distances. CLI Prompt Menus One-line CLI Prompt menus occupy the SVL Command Line at the top of the window Press (Esc) to exit the prompts or choose to delete the process using the ‘Cancel’ button on the top right. MOE Selection Menu Extend selection set Deselect all atoms Invert current selection Knowledge-based selectors for different parts of protein/ligand bound structures Atom Selection Tool for advanced selecting Save and Restore selection sets Selection state of residues is coordinated with the Sequence Editor Pull down menus for selection by property, element, extension or other criteria The Atom Selector MOE | Selection | Atom Selector Selection Restrictions General Selection actions Logic Operations Save and Load selection sets; create named sets Select by elements and atom types Select by name Select by SMILES string substructure Other Selection Options: 1. Accessibility 2. Chirality 3. Connectivity 4. Geometry 5. General 6. Pharmacophore 7. Protein (e.g. alpha carbon) Extend Selection Criteria Moving Atoms with the Middle Mouse Button XY Rotate Middle Drag: XY Translate <Shift> Middle Drag: Zoom in/out <Ctrl> Middle Drag: center of rotation Middle click to center on atom <Alt> Middle Drag: XY Rotate selected only <Alt> <Shift>Middle Drag: XY Translate selected only Exercise: Moving Atoms with the Middle Mouse Button 1. View the coordinate system (Render | Draw | Coordinate Axes). 2. Rotate view about the XY axes (Middle Drag). 3. Translate view (<Shift>-Middle Drag). 4. Middle click on the carbonyl O to move the center of rotation. 5. Remove the coordinate axes by de-selecting (Render | Draw | Coordinate Axes) 6. Deselect all atoms (Left-click in space or (Selection | Clear)). 7. Move a selected subset with <Alt>-Middle Drag (rotate) <Shift><Alt>Middle Drag (translate). MOE Render Menu Center, Save and Load views Draw H-bonds, VDW contacts, label options, coordinate axes, etc. Stereo viewing options: Quad-Buffer, OverUnder, Interlace, Left-Right, Parallel Protein/DNA backbone rendering Atomic/Molecule object rendering Hide and Show various sets Basic coloring Atom Labeling menu Detailed atom and label style menu Setup of default colors and object dimensions. Exercise: Small Molecule Rendering • Rendering actions apply to: All atoms (if none are selected) Selected atoms only (if there are selected atoms) Select and render as ‘Space Filling’ Select group and render as ‘Stick’ 1. Deselect all atoms. 2. Use Left drag click to select part of the molecule. Render it as space filling (Render | Space Filling). 3. Select other parts of the molecule using left-drag or other methods, and render them as stick, ball and stick, or line. Protein Backbone Rendering MOE | Render | Backbone or MOE | Popup | Backbone Turn off backbone Various Backbone rendering styles Backbone coloring options Exercise: Open PDB file prior to rendering complex 1. Close the current system: MOE | File | Close 2. Select the file MOE | File | Open $MOE/sample/mol/1pph.pdb. 3. Press ‘Load PDB File’. 4. A variety of options are available in the PDB File panel. Choose to centre the view and press ‘OK’. Exercise: Rendering Trypsin with Ligand 5. Select the water molecules (Selection | Solvent) and delete them (RHS | Delete). 6. Select the ligand(s) (Selection | Ligand) and render as space filling (Render | Space Filling). 7. Use Render | Color to select a desired colour for the selected atoms (green). 8. Deselect the atoms 9. Draw a backbone ribbon through the selected atoms (Render | Backbone | Slab Ribbon). 10. Color the backbone by Chain Color (Render | Backbone | Color | Chain Color). 11. Hide the selected protein atoms (Render | Hide | Receptor). 12. Click on empty space to clear selection state. Exercise: Protein-Ligand Pocket Rendering 1. Turn the backbone off (MOE|Popup|Backbone|None) 2. Show ligand and pocket (MOE|Popup|Show|Ligand, Pocket) 3. Label the residues (MOE | RHS | Label | Residue) 4. Draw H-bonds (Render | Draw | Hydrogen Bonds) 5. Center the image (RHS | View) which should now look like the image on the left. 6. Save the system as a MOE file, MOE | File | Save trypsin_pocket.moe Contact Statistics Calculate and display probability of finding hydrophobic or hydrophilic contact at a point P relative to an atom. The contacts are derived from PDB x-ray structure statistics. Preference for Hydrophilic Contacts r P v u Preference for Hydrophobic Contacts Contact Statistics (cont.) Contact Statistics can be used to highlight directional packing preferences on interaction surfaces: Hydrophilic contacts for polar H Hydrophobic contacts above and below pi system Contacts statistics on top of interaction surface Interaction Surface Exercise: Contact Statistics in Pocket 1. Open the Contact Statistics panel: MOE | Compute | Surfaces and Maps 2. Setup the panel as follows Surface: Contact Preference 3. Press Apply. 5. Save to a MOE file (File | Save) ‘trypsin_csats.moe’ toggling on Graphics: All in panel Exercise: Receptor Molecular Surfaces 1. Close the current system. (File | Close). 2. Disable hydrogen bond selection by de-selecting MOE | Render | Draw | H bonds 3. Open the files (MOE | File | Open ) $MOE/sample/mol/biotin.moe. $MOE/sample/mol/biotin_rec.moe. 4. Calculate partial charge (MOE | Compute | Partial Charge) and enable “Adjust Hydrogens and Lone Pairs as Required” 5. Draw a electrostatic surface about the pocket (MOE|Compute|Surfaces and Maps) 6. Name the surface ‘Pocket Surface’ 7. Color by Electrostatics 8. Press Apply Exercise: Receptor Molecular Surfaces 1. To isolate the pocket atoms, press Isolate on the panel 2. Turn off backbone (MOE|Popup|Backbone|None) 3. Select the pocket atoms (MOE|Popup|Select|Pocket) 4. Label the residues with the residue name (MOE|RHS|Label|Residue) Exercise: Ligand Molecular Surfaces 1. Now draw a molecular surface of the ligand (MOE | Compute | Surfaces and Maps) selecting the defaults, but changing the Name to: Ligand Surface, and selecting Atoms: Ligand Atoms 2. Press Apply Exercise: Biotin receptor surface (cont.) To view the different surfaces, go to (MOE | Window | Graphic Object) Select Pocket Surface Press Hide Select both surfaces (Shift Left mouse click) Press Toggle to switch between surfaces Surfaces: Backface Culling and Visualization Note the transparency options for the front (TF), and the back (TB) Set the slide on TB, and rotate the system to view the backface culling Ligand Interactions • Automatic 2D protein-ligand interaction diagrams – Application of MOE's automatic 2D depiction algorithm – Easily identify polar, hydrophobic, acidic and basic residues – Visualize solvent exposed ligand atoms and residues – Visualize sidechain and backbone acceptor and donor interactions • Visualize 3D Contacts – Display hydrogen bonds between ligand, receptor/solvent and metal ligation – Score estimates strength of hydrogen bond • Report protein-ligand interaction data – Textual listing of interactions with scores • Export 2D schematic to a picture – Choose between png, gif, jpeg, bmp and copy to clipboard Exercise: 2D Protein-Ligand Interactions 1. Hide all surfaces (MOE | Window | Graphic Object). Select and Hide each surface 2. Open MOE | Compute | Ligand Interactions Exercise: Protein-Ligand Interactions acidic residue amount of ligand contact polar residue substitution contour solvent exposure sidechain donor backbone donor/acceptor greasy residue Exercise: Protein-Ligand Interactions 1. In the Ligand Interactions panel, select 3D Contact Style 2. Turn ON Residue H-bond Distance. Residue hydrogen bonds are scored and distance metrics are drawn in the main MOE window In the main MOE window, observe the relative strength of the ideal hydrogen geometry, shown as dotted lines 2. The Sequence Editor The SE Displays a ‘2D’ view of the molecular data Open SE… (MOE | SEQ) or <Ctrl>-Q MOE Window - 3D molecular data is displayed in the Sequence Editor as 2D data: bound ligand(s) protein chain(s) water chain(s) Objects in SE can be used to manipulate objects in MOE Window (ensure Selection | Synchronize is enabled) Secondary Structure in SE is displayed as colored bars Anatomy of the Sequence Editor (SE) SE Menu Alignment Ruler Chain Label Residues Chain Number Footer Secondary Structure Bars Red = helices Yellow = sheets Blue/Green = H-bonded Turns SE Menu Command Conventions • Commands from the Sequence Editor are preceded by SE. (SE | Selection | Residue Selector) Synchronize selection of objects in MOE Window or DBV (via MOE or SE | Selection | Synchronize) Data Hierarchy in MOE Exercise: Molecular Hierarchy 1. For the loaded biotin-streptavidin system, toggle off the molecular surfaces, at MOE | Windows | Graphic Objects. Select the surface and press Hide. 2. Show receptor, using MOE | Render | Show | Receptor. 3. Clear Labels (MOE | RHS | Label | Clear) 4. Open the Sequence Editor (MOE | SEQ). The system should appear as shown: 4. Click in empty space to clear selection state Exercise: Molecular Hierarchy (cont.) 5. Color the atoms by chain color (MOE | Render | Color | Chain) 6. Turn on the compound names (SE | Display | Compound Name) 7. Turn on the secondary structure color bars (SE | Display | Actual Secondary Structure). Selecting Objects in the Sequence Editor Select residues one at a time Select Multiple Residues Select range of residues <Shift> <Ctrl> Chain Selection: Click: Chains one at a time <Ctrl> Multiple Chains <Shift> a range of chains Left Drag: Selection Box Sequence Editor Popup Menus Open the SE Popup menus by right-clicking over the areas shown below: Exercise: Using SE for Protein Rendering 1. Select protein chain (chain 2). Position mouse over chain and use Right mouse button to get Chain popup. 2. Select Backbone | Slab Ribbon, and 3. Backbone |Color | Chain color 4. Hide receptor (Atoms | Hide) 5. Select Chain 1 (ligand) and use popup menu to Render | Space Filling Exercise: Using SE for Protein Rendering (cont.) 6. Close the current system (MOE | RHS | Close) 7. Close all windows except the main MOE window 3. The MOE Database Viewer Molecular Data easily transferred between database and MOE Window Character, numeric and molecular data fields Full 3D molecular structure Anatomy of the Database Viewer (DBV) Menu Bar Field Headers DBV CLI Entry Numbers Data Cells DBV Menu Command Conventions • Commands from the Database Viewer preceded by DBV (DBV | Entry | Show All Entries) DBV Left Mouse Button Commands Select entries/fields one at a time Select multiple entries/fields Select range of entries/fields <Ctrl> <Shift> DBV Popup Menus The Popup Menus are invoked with the right mouse button Exercise: Opening a MOE Database Viewer 1. (File | Open) Select the file $MOE/sample/mol/opiates_analog.mdb. 2. Open in a database viewer (Open in Database Viewer). Exercise: Opening a MOE Database Viewer (cont.) 3. Left-Diagonal drag on a molecule cell to enlarge it. 4. Middle-drag in the molecule cell to rotate the view. XY Rotate Enlarge Molecule View: Left Diagonal Drag on Molecule Cell Middle Drag: Zoom in/out <Ctrl> Middle Drag: Exercise: Database Printing and Tiling 1. (DBV | File | Print) 2. Click on ‘Tile Molecule Field’. 3. Select ‘Display Entry Number’ and choose the footer to be the field ‘name’. 4. Change Grid: 3x4 Exercise: Copying Morphine from the DBV 1. Close the current system in the MOE Window. 2. Copy morphine (entry 1) to the main MOE window by Left-doublemouse click in the mol field 3. Select ‘Clear Molecular Data’ 4. Render as stick (MOE | Render | Stick) Exercise: Protonate the nitrogen atom in morphine 1. Left-click on the nitrogen atom, so that it becomes highlighted. 2. Left-click on the ‘Builder’ button on the RHS of the menu bar. 3. Select +1 for the ionisation state. 4. The nitrogen atom is then protonated. Molecular Mechanics • Aims to predict the structure and properties of molecules. • Uses a Force Field with parameters from known structures • Energy Minimization calculates the energy of a molecule and adjusts the structure to obtain a lower energy structure. • Predicting short-range steric interactions is easy and accurate • Predicting long-range electrostatic interactions and the effect of water is difficult. • Flexible molecules may need to be described with an ensemble of conformations. Potential Energy in MOE E = ESTR + EANG + ESTB+ ETOR + EOOP + EELE + EVDW + ESOL Forcefield title Toggle on/off terms in the potential Adjust non-bonded interaction switching function No. of parallel processor threads to be used Forcefield parameter file Load different forcefields Adjust electrostatics implementation Partial charge calculation according to selected potential Supported Forcefields Biopolymers (proteins and nucleic acids) AMBER 89, AMBER 94, AMBER 99, CHARMM 22, CHARMM 27, OPLS-AA Small Molecules MMFF94, MMFF94s, MMFF94x Crystallographic Engh-Huber Carbohydrate PEF95SAC Simple Molecular Modelling Rule Exercise: Forcefield Energy Minimizations (1) 1. First choose an appropriate potential and partial charges in MOE | Window | Potential Setup Click on arrow by Load Select the MMFF94 potential Select ‘Fix Charges’ to assign atomic charges according to the chosen potential 2. Press OK and Close Exercise: Forcefield Energy Minimizations (2) 2. Choose MOE | Compute | Potential Energy System energy components Potential energy components are also shown in the SVL window Exercise: Forcefield Energy Minimizations (3) • Minimizations may be forcefield, or semi-empirical (MOPAC 7) Hamiltonian based Automatically add H’s (and LPs if required) Automatically assign partial charges Force current (R/S) stereochemistry Tether Weight (kcal/mol A2) Potential Setup window Exercise: Forcefield Energy Minimizations (4) To minimize the molecule, select (MOE | Compute | Energy Minimize) 3. Use the defaults in the panel and press OK Minimized Morphine Exercise: MOPAC Minimization Select (MOE | Compute | Energy Minimize) PM3, AM1 or MNDO To view HOMO/LUMO orbitals go to (MOE | Window | Graphic Objects) Option to plot and view orbitals (HOMO and LUMO) Calculating Interaction Potential Energies 1. Close the current system (RHS | Close) 2. Open biotin and its receptor (MOE | File | Open ‘$MOE/sample/mol/biotin.moe and biotin_rec.moe’ 3. Add Hydrogen atoms and compute partial charges (MOE | Compute | Partial charge) 4. Select the ligand. Right click in the main MOE window to get popup panel. Popup | Select | Ligand 5. Choose MOE | Compute | Potential Energy ALL: total system E SEL: selected only E INT: selected – unselected interaction E Exercise: Dihedral Energy Plots • Plots the energy about a single rotatable bond. 1. Close current system. Open $MOE/sample/mol/biotin.moe 2. Add hydrogen atoms (MOE | Edit | Hydrogens | Add Hydrogens) 3. Open the dihedral energy plot panel: MOE | Compute | Mechanics | Dihedral Energy Plot. 4. Select four consecutive carbon atoms in a dihedral. Exercise: Dihedral Contours • Plots the energy contours about two rotatable bond. 1. Open the Dihedral Contour prompt (MOE | Compute | Mechanics | Dihedral Contour Plot). 2. Select four consecutive carbon atoms in one dihedral, followed by four consecutive carbon atoms in another dihedral. Forcefield Restraints: Energy terms • The restraint energy is a sum of all the individual restraints: ERESTRAINT = S EDistance + S EAngle + S ETorsion ETorsion EAngle EDistance • When restraints are set, their energy and forces are included in ALL MM based calculations. Creating Forcefield Restraints • Restraints are created from the MOE | Edit | Potential | Restrain command. • The type of restraint and the parameters are set in the following CLI prompters. • ‘Create’ must be pressed to create the restraint. EDistance = ( max (0, L2 - r2)3 + max (0, r2 - U2)3 ) * w EAngle = (max(0, cos a - cos L)3 + max(0, cos U - cos a)3 ) * 100 w ETorsion= ( (1 - cos max(0,d - L))3 + (1 - cos max(0,U - d))3 * 10000w Exercise: Creating Forcefield Restraints 1. To create a distance restraint open (MOE | Edit | Potential | Restraint). Select the acid oxygen and a hydrogen alpha to it. Set the Target Limits as (L = 3.0, U = 3.5, w = 1). Press Create. 2. Similarly, create an angle restraint (L = 1150, U = 1350 , w = 1) between the carboxylate C and the O and H atoms shown here. Minimized with restraints 3. Minimize the structure (Compute | Energy Minimize). The Tethers and Restraints Panel The Tethers and Restraints panel (Window | Potential Setup | Restraints) can be used to manage and edit current restraints. Toggle ‘Restraints’ to display restraints List of current restraints Edit selected restraint. Press ‘Apply’ to institute changes. Delete selected restraints Exercise: Removing Restraints 1. Open the Tethers and Restraints panel (Window | Potential Setup | Restraints). 2. Delete all the current distance and angle restraints. 3. Re-minimize the molecule (Compute | Energy Minimize). Minimized with restraints Minimized without restraints Exercise: Using the GizMOE Minimizer The GizMOE Minimizer is a minimizer that runs continuously in the background. 1. With biotin in the system, start the GizMOE Minimizer. MOE | GizMOE | Minimizer 2. Left drag to select and move part of the molecule. Then watch how the energy and geometry are automatically updated. <Alt> <Shift> Drag Translate Selected Atoms Only 3. Turn off the GizMOE Minimizer. Click the Cancel button and choose GizMOE_Minimizer[]. If necessary re-minimize the system (RHS | Minimize) Conformational Searching • Generation of different conformations of a molecule or a complex is very useful for drug design. • Conformational search methods available in MOE Systematic Conformational Search Stochastic Conformational Search Conformational Database Import Molecular Dynamics Stochastic Conformational Search • Random sampling of local minima on the potential energy surface E 1. Perturb geometry Energy Cutoff 2. Minimize E0 Torsion Space Stochastic Conformational Search Panel Conformation Generation: Output database Randomly: Invert chiral centers Rotate torsions Perturb xyz coordinates Conformation Minimization: Systematic Conformational Search • Exhaustive incremental dihedral rotation search E Cutoff E0 Torsion Space Systematic Conformational Search Panel Add/Remove dihedrals from list Set dihedral increment Output Database Minimise structures List of bonds to undergo rotation Exercise: Systematic Conformational Searching (1) 1. Close the current system (RHS | Close) 2. Open up the MOE file for the molecule built earlier (MOE | File | Open ‘my_first_molecule.moe’). 3. Perform a systematic search on this molecule using the default options (MOE | Compute | Conformations | Systematic Search) 4. Left-Drag in DBV molecule cell to view structures. Enlarge Molecule View: Left Diagonal Drag on Molecule Cell Exercise: Systematic Conformational Searching (2) 1. Open (DBV | Compute | Descriptors). 2. Enter ‘Energy’in the Filter field. 3. Select the descriptor (Left mouse click once) “E Potential Energy” and press OK. Exercise: Sorting and Selecting Conformers 1. Position the mouse over the E Field. Right click to use Field Header popup to Sort UP on energy. 2. Left double click on the lowest energy conformer in the mol field to copy to the MOE Window. Superposing Conformations Database to perform calculations on Mol field to perform calculations on Measurements to perform on database Superposition of conformers in database Auto-Label atoms by element and number Exercise: Superposing Conformations 1. Left mouse drag to select the methyl substituted pyridine ring 2. Bring up the Conformation Geometries panel. (DBV | Compute | Conformation Geometry…) 3. Change Molecule Field: to Overwrite Current Field. 4. Click on the Selected Atoms buttons. 5. Click on the Superpose button. Exercise: Superposing Conformations (cont.) 6. Shift Left mouse click over a subset of entries (try entries 1 to 5) 7. Use Molecule Cell popup to Copy Selected Entries to MOE Window 8. Observed the superposed conformations 9. Color by chain using (MOE | Popup | Color | Chain) Diverse Conformational Subset 1. Open the Diverse Subset panel (DBV | Compute | Diverse Subset). 2. Set the Output Limit to 20. 3. Choose ‘Conformation’ as the selection method. 4. Press OK to start calculation. Exercise: Diverse Conformers Subsets 5. Use Field popup to Sort Up on $DIVPRIO. 6. Copy 20 diverse conformers to MOE with popup. Shift Left click over entries 1 to 20. 7. Position mouse in mol field and use Right mouse button to get Popup. Select Copy Selected to MOE 8. Remember to select Clear Molecular Data 9. Render conformers as stick (MOE | Render | Stick) Interactive Superposition • Edit | Interactive Superpose is a tool for optimally superposing molecules based on selected point sets. • More than two structures may be superposed simultaneously. Exercise: Interactive Superpose (1) 1. Close the current system and open (File | Open) $MOE/sample/mol/opiate_analogs.md b 1 2 3 2 1 2. Select entry 1 and 7 (morphine and heroin). Copy to MOE window 3 3. If molecules are superposed, separate by CtlLeft click on an atom of one molecule, to select entire molecule. 4. Separate by moving selected molecule using Shift-Alt-Middle mouse 5. Center the view (RHS | View). 6. Render the structures as ball and stick (Render | Ball and Stick). 7. Hide the hydrogens (Render | Hide | Hydrogens). 8. Initiate superpose (Edit | Superpose). Exercise: Interactive Superpose (2) 6. For Set 1 select the indicated oxygens labelled (1) on each molecule 7. Press Set: 2 in the CLI prompt and select the indicated aromatic ring carbons labelled (2) 8. Press Set: 3 in the CLI prompt and select the indicated oxygen atoms directly connected to the benzenes labelled (3) 9. With the minimum 3 point sets specified, the Superpose is possible. Press Superpose to superpose the structures. 10. Pressing Superpose will superpose the structures based on an optimal RMSD. Flexible Alignment of Small Molecules - Feature-based alignment of 2 or more molecules - Features are pharmacophore-like - Stochastic search algorithm employed for flexibility - Weighting scheme for features Exercise: Flexible Alignment of Opiates (1) 1. 2. 3. Close the current system (RHS | Close) and import morphine, heroin and demerol (entries 1, 7, 11) from the database $MOE/sample/mol/opiate_analogs.mdb Ensure that the partial charges have been set, using MOE | Compute | Partial Charges. Select one of the molecules using Ctl-Left mouse click on an atom of one molecule and fix it: MOE | Edit | Potential | Fix. Exercise: Flexible Alignment of Opiates (2) 6. Choose MOE | Compute | Conformations| Flexible Alignment. Decrease the iteration limit down to 20, instead of 200. 7. Preserve defaults and press OK Similarity terms and weighting Exercise: Flexible Alignment of Opiates (3) 8. Let the application run to completion. 9. Sorting in S occurs automatically 10. Choose the “best” alignment “Best” may be that with the lowest scoring function value – but take strain into account! Exercise: Flexible Alignment of Opiates (4) 11. Copy the “best” alignment into the MOE Window. - Aligning multiple molecules can be time-consuming; try aligning them one at a time, keeping the earlier alignments fixed. Exercise: Rendering of the Flexible Alignment Select MOE | Render | Color | Chain. This will colour the chains (i.e. separate molecules) of the flexible alignment. Close the current system (RHS | Close) Close all windows except the main MOE window Further Simulation Techniques • Poisson-Boltzmann electrostatics e.g. analysis of active site in a receptor can reveal the effect of the surrounding residues on the binding properties of a ligand. Solution of the full non-linear PB equation, allowing for different ion classes, radii and partial charges. • Molecular Dynamics e.g. use to relax structures and to generate conformational states at a desired temperature and/or pressure (in NPT, NVT, NVE, NPH). Further Simulation Techniques • Docking • Flexible ligands and a rigid receptor. The poses may be constrained to fit a pharmacophore query. • Affinity dG scoring is used to estimate the enthalpic contribution to the binding free energy of hydrogen bonding, ionic, metal ligation and hydrophobic interactions. Introduction to Database Viewer Analysis Used for: •Cheminformatics •QSAR •Clustering •Similarity Search •Diverse Subsets •Fingerprints •Library Generation/Design •Ph4 applications •Output for •Conformation search •Dynamics •Flexible alignment •Docking •Washing / Processing Exercise: Opening a MOE Database Viewer 1. (File | Open) Select the file $MOE/sample/mol/blood_brain.mdb. 2. Open in a database viewer (Open in Database Viewer). 3. Save a local copy (DBV | File | Save ‘bbb.mdb’) Exercise: Calculating Descriptors 1. Open the QuaSAR-Descriptor panel (DBV | Compute | Descriptors). 2. On the Filter line, type TPSA. Left click once on TPSA in the panel to select. 3. Repeat to select Weight, logP(o/w), and MR 4. Press OK and descriptors will be calculated into the database Descriptor Filter Exercise: Sort by Activity • Sort in descending order of logBB 1. Open the Sort Database panel (DBV | Compute | Sort). 2. Select Field: “logBB” 3. Enable “Descending” 4. Press OK Exercise: Plotting Data 1. Open the DBV Plot window (DBV | Display | Plot). 2. Select logBB as the numeric value to plot. 3. Use the Right button in the plot area to compute the range with the DBV Plot popup. Mouse Actions in the DBV Plot Window Entry Selection is reflected in the DBV and the DBV Plot window Drag on axis: Selection Range Left Click: Select points individually <Shift> Drag: <Ctrl> Drag: XY Translate Plot Zoom in/out of Plot Drag: Selection Box Exercise: Select actives • Select compounds with logBB > 0 1. Select active compounds by using Left mouse drag in Plot:Display for all entries where logBB > 0. 2. Notice selected entries are updated automatically in the database viewer Exercise: Hide Inactives • Hide all compounds with logBB < 0 1. Since all compounds with logBB > 0 are selected, go to (DBV | Entry | Hide Unselected entries) 2. Use the Right button in the plot area to compute the range Exercise: Look at active compounds 1. Launch database browser by going to (DBV | File | Browser) 2. Select Subject:mol (2D) for depicted mode 3. Use forward/backward triangles to navigate Exercise: Plot descriptor and activity relationship 1. Show all entries (DBV | Entry | Show All Entries). 2. Start the database correlation plot prompt (DBV | Compute | Analysis | Correlation Plot…). 3. Pick ‘TPSA’ and ‘logBB’ to plot along X and Y. Exercise: Show relationship between all fields 1. Start the database correlation matrix prompt (DBV | Compute | Analysis | Correlation Matrix…). 2. Press on TPSA/logBB to get same correlation plot Exercise: Select actives Select points in the plot: Drag: Selection Box Entry Selection is reflected in both the DBV and Correlation Plot Use the Attributes menu to change look of the plot Exercise: Show relationship of actives with logP(o/w) 1. Hide inactives, go to (DBV | Entry | Hide Unselected entries) 2. Start the database correlation plot prompt (DBV | Compute | Analysis | Correlation Matrix…). 3. Press on logP(o/w) / logBB to get correlation plot Exercise: Show clustering of actives and inactives 1. 2. 3. 4. 5. 6. 7. Show all entries (DBV | Entry | Show All Entries) Open 3D Plot (DBV | Compute | Analysis | 3D Plot) Set X to “Weight”, Y to “TPSA”, Z to “logP(o/w)” Set activity to “logBB” Set Threshold to 0 Press Plot Enlarge points using (MOE | Render | Ball and Line) Pharmacophore Overview Aim: to find chemically unrelated molecules which share molecular features 1. Take an active molecule. 2. Annotate possible PH4 HB Acceptor features Aromatic 3. Take conformations of a set of diverse molecules. 4. Annotate with PH4 features 5. Find hits which match the query. HB Acceptor 3. Create a query with these features Compute | Conformations | Pharmacophore Elucidation Objective Starting from single conformations of active and inactive compounds, sample conformations on the fly and automatically extract maximum common Ph4 pattern which selectively recognizes active features. Output database Activity threshold: binary or no activity Selection of Ph4 schemes that can be stored and loaded Text report Ligand database Specification of conformational method Feature list and feature properties Modification of features / rules Parameters for structure alignment Pharmacophore search parameters Exercise: Pharmacophore Elucidation I The Elucidator will try to identify popular Ph4 patterns from sets of unaligned molecules. To validate the performance of the Elucidator, we will start with an example where we know the “optimal” result (aligned by nature in X-ray protein structures): 1. 2. 3. 4. 5. Open Elucidator panel in (MOE | Compute | Conformations | Pharmacophore Elucidator) Choose an output database name (default: ph4elucidate.mdb) Browse to select as Input Database: $MOE/sample/mol/1RO6_ligands.mdb This has 7 ligands from pdb structures Switch the Conformations setting to Bond Rotation. Leave the Activity Field as “All Active” since all ligands are active in this example (otherwise you would select the activity/inactivity threshold here) Remain with the default Ph4 scheme (CHD) and click OK. Pharmacophore Elucidation II The output database looks like… Conformations of Ph4 alignment Query features: D/A = heavy atom Don/Acc d/a = projected Don/Acc H = Hyd/Aro m = Metal +/- = Cation/Anion Active molecules Separation of actives/inactives Accuracy of actives Query information for DB Browser Alignment score Probability by chance Accuracy of inactives Number of features of specific type Total Number of features Exercise: Pharmacophore Elucidation III The output database is sorted by ascending overlap (alignment) score. 6. Use (DBV | File | Browser) to examine each Ph4 alignment. Note the modified view of the browser while displaying the results. You may want to modify input parameters in your elucidator calculation interface if you are not satisfied with the quality of the results or you may directly edit the underlying queries to further refine the results. You may want to save the current Ph4 query or modify the features of a given entry. Double-clicking in the query cell in the Database Viewer will launch the Ph4 Query Editor. Edit in the Database Browser brings up the Ph4 Query Editor. 4. The SVL Commands Window SVL is a powerful language designed to allow you to customize MOE and extend MOE with your own functions 1. Open the SVL Commands Window with (RHS | SVL) Exercise: SVL Commands Window 1. SVL commands are prefixed in the text with svl>. For example, enter 3+4 in the SVL Commands Window: svl> 3+4 7 Press Enter 2. SVL commands can be used to open menus and build molecules from SMILES strings For example, build methane by entering svl> sm_Build ‘C’ Basic SVL windows in MOE Text Editor (TED) Modules & Tasks Manager ASCII file / SVL program editor Program control / Source Code Crash History Source-level error trace-back Appendices Forcefield File alkane.ff: Atom Typing Block #moe:forcefield 2000.02 #comment lines title ALKANE disable oop stb itortype CT C 'sp3 C'type HC H 'H attached to alphatic C'[rules] #--------TYPE ASSIGNMENT RULES -------- CT match '[CX4]‘ HC match '[#1]C‘ … Forcefield File alkane.ff: Bond Stretch Block ESTR wSTR K 2ij (rij Lij ) K 3ij (rij Lij ) K 4ij (rij Lij ) 2 3 4 ij [str] #--------------------- BOND STRETCH -------------------- #code T1 T2 LEN K2 K3 K4 bci #----- ---- ---- ---- ---- ---- ----- ---- * CT CT 1.518 448.2 -1010.6 1329.08 - * CT HC 1.090 334.2 -606.37 641.608 - Forcefield File alkane.ff: Angle Bend Block E ANG wANG K 2ij ( O ) K 3ij ( O ) K 4ij ( O ) 2 3 4 ijk [ang] #------------------ ANGLE BEND --------------------ang-function angle #CODE T1 T2 T3 ANG K2 K3 K4 #--- ---- ---- ---- ----- ----- --- ----- * CT CT CT 109.50 86.97 0.00 0.00 * CT CT HC 109.50 87.15 0.00 0.00 * HC CT HC 109.50 74.07 0.00 0.00 Forcefield File alkane.ff: Torsion Block 5 ETOR wTOR K N ;ijkl 1 cos(Nijkl ) ijkl N 1 [ptor] # ------------- proper torsion ---------------------# T1 T2 T3 T4 V1/2 V2/2 V3/2 V4/2 V5/2 # -- -- -- -- ---- ---- ----- ---- ---- * CT CT CT CT 0.00 0.00 1.606 0.00 0.00 1 * CT CT CT HC 0.00 0.00 0.250 0.00 0.00 1 * HC CT CT HC 0.00 0.00 0.221 0.00 0.00 1 Forcefield File alkane.ff: Electrostatics Block E ELE wELE 5 qi q j e2 d 4 0 i j N 1 (rij bELE ) k [nonbonded] # ------ nonbonded information ---------ele-dielectric 1 # dielectric+distance dependent flag ele-buffer 0 # electrostatic buffering ele-scale14 1 # 1-4 interaction scaling ele-charge-fcn alkane # svl fcn to compute charges Forcefield File alkane.ff: VDW Block EVDW (1 a) Rij wVDW ij i j rij aRij vdw-scale14 nij nij (1 b) Rijmij mij nij mij mij mij mij (rij bRij ) 1 [vdw] # ------- VDW PARAMTERS ---#T1 T2 R EPS m n #--- ---- ------- ------- -- -- CT CT 3.6458 0.21949 12 6 CT HC 3.5834 0.01799 12 6 HC HC 3.5220 0.001475 12 6 Visualization Setup: Coloring MOE | Render | Setup… Set colors of objects Press Apply to institute changes Restore defaults Save new settings as defaults Visualization Setup: Dimensions Protein Ribbon dimensions Atom and Bond dimensions Visualization Setup: Lighting and Projection SVL and MOE-batch • MOE/batch • Terminal-style interface (no GUI). • SVL commands entered at prompt. • Used for scripting long tasks and automating procedures.