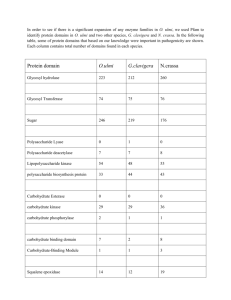
activity
advertisement

Activation of biomolecules
Activation of small biomolecules
Activation of small inorganic biomolecules in order to
make them reactive is necessary both for aerobic and
anaerobic organisms.
These reactions provide the necessary energy for their
life.
Aerobic organisms: O2 (to water), N2 (to NH3), H2O and
CO2 (photosynthesis) activation
Anaerobic organisms: H2, CO, CO2, CH4 activation
The reactions are catalysed by metal ions with variable
oxidation states: Fe, Cu, Mo, Mn, V, Ni containing
metalloenzymes.
Triptophane dioxygenase
H2
C
CH
N
H
O
COOH
H2
C
CH
COOH
18
+ O2
NH2
NH2
NH
HC
18
O
In the resting state the enzyme hem contains a high spin FeII and the
coordination position 6 is empty.
When the substrate is bound to the enzyme conformation changes
and it will be able for reversible O2 binding,
[SFeO2] transition complex is formed, in which oxygen is in the form of
O2•-.
After an oxygen insertion step of FeIII- O2•- with the double bound
substrate, a rearrangement and finally a cleavage of the bonds occur.
Cytochrome P450
The electrontransfer component of the monooxygenase enzymes.
R−H + O2 + 2 e− + 2 H+ → R−OH + H2O
Solubilisation of compounds containing C-H bonds,
Metabolism of lipids and other compounds
Nomenclature: λmax (CO adduct) = 450 nm (instead of the usual 420 nm)
Structure: M ~ 45.000
Resting state: FeIII (low spin)
N = 5 (Cys-S axial coordination)
(coordination site 6: labile water molecule → oxygen
binding site)
Mechanism: FeIII−OH2 (ε=−300 mV) → FeIII, R−H (ε=−173 mV) →
FeII, R−H → FeIII−O2−, R−H → FeIII−O22−,R−H →
FeVO, R−H (vagy FeIVOP·+) → FeIII−OH2 + R−OH
Cytochrome P450
The Figure depicts the
adduct formed with
thio-camphor
It catalyses
hydroxylation
of thio-camphor.
The catalytic cycle
of cytochrome P450
The key steps:
Formation of the
reactive oxenoid
oxoferryl(V) (= FeVO)
or
oxoferryl(IV)porphyrin-radical
(= FeIVO−P·+).
A comparative table of the iron and copper
containing proteins
Function
O2 transport
oxygenase function
oxidase inhibition
electron transport
antioxidative function
NO2– reductase
Fe protein
(h: heme protein)
(nh: non-heme protein)
hemoglobin (h)
hemoerythrin (nh)
cytochrome P-450 (h)
methane monooxygenase (nh)
pyrocatechin dioxygenase (nh)
peroxidases (h)
peroxidases (nh)
cytochromes (h)
peroxidases (h)
bacterial superoxide dismutase (nh)
Cu protein
heme containing nitrite reductase (h)
Cu-containing nitrite reductase
hemocyanin
tyrosinase
quercetinase (dioxygenase)
amine oxidase
laccase
blue Cu proteins
superoxide dismutase (Cu, Zn)
Copper states in proteins
Type I: Blue copper proteins
Resting state: Cu(II), paramagnetic,
unusual vis and EPR parameters
ε ~ 100 εnormál
A║ << Anormal
Type II: Non-blue copper proteins
Spectral parameters characteristic of the tetragonally
distorted Cu(II) complexes (light blue proteins)
Type III: EPR inactive copper proteins
- Cu(II) dimers (antiferromágnetic coupling)
- Cu(I) state
CuA: Mixed valence copper proteins
Cu(I) - Cu(II) pair
More familiar copper proteins
Name
Total
Plasstocyanin
Azurin
Stellacyanin
I.
II.
III.
Function
1
1
1
1
1
1
-
-
electrontransfer
2
100%
enzyme
Cu storage
detoxification
100%
O2-transport
Superoxidedismutase
Metallothioneine
2
-
1-10
-
Hemocyanin
>10
-
-
Tyrosinase
4
-
-
4
2
4
1
1
1
4
Ceruloplasmin
Laccase
Ascorbic acid
oxidase
6
4
8
3
oxygenase
2+1
2
Cu-transport
oxidase
Blue copper proteins
Occur mostly in plants (preparation from algae)
Have important role in photosynthesis as electron transfer proteins.
Characteristics:
- low molecular mass (M ~ 10 000, ~ 100 am acid + 1 CuII)
- Intense blue color
λ ~ 600 nm,
ε ~ 3000 - 5000
- EPR activ, low A|| coupling constant
- high redox potential (ε ~ + 0.3-0.7 V)
(easy reduction)
Mechanism:
Cu(II) - SR
Cu(I) + .SR
fast reaction
Structure:
Cu(II) in unusual environment distorted tethedron
(usually:
2 his +1 cys + 1 met)
Non-blue copper proteins
Characteristics: paramagnetic, ESR activ
pale blue ( 10 -100)
Cu(II), d9
Structure/bonding:
- tetragonally distorted octahedron
(like Cu(H2O)62+ or CuL4(H2O)2)
- there is one labile ligand in the coord. sphere
(e.g. a water molecule in axial or equatorial position
substrate binding site
Occurrance:
CuZn-SOD
Non-blue oxidases (pl. galactose oxidase, amin oxidase)
Blue copper oxidases (I + II + 2 III)
ESR inactiv copper proteins
Structural characteristics:
In the resting state they contain either 2 close, but independent
CuI-ions, or 2 antiferromegnetically coupled CuII-ions.
In the coordination sphere of Cu there are usually 3 N(His)
donor atoms, while at the fourth position the substrate/O2 is
bound.
Occurrance:
hemocyanin: oxygen carrier
enzymes: tyrosinase (mixed monooxygenase/oxidase function)
blue copper oxidases:
e.g. ceruloplasmin, laccase, ascorbic acid oxidase, etc.
occurs with type 1 and 2 copper
(type 2 and 3 form a trimer)
Tyrosinase enzyme I.
Structure: Similar to hemocyanin but it contains only two
subunits
(= 2 + 2 copper).
CuII−O22−−CuII
reversible oxygen transfer but in enzymatic reaction.
The enzyme acts in a mixed function catalytic reaction:
- monophenolase (monooxygenase) activity
- diphenolase (oxidase) activity
The different function from hemocyanin can be explained by the
different protein character.
Action: 2 CuI + O2
Similar structure, but different function occurs in the groups of iron
proteins: e.g.. hemerythrin (O2 transport) and
methane monooxigenase or ribonucleotide reductase
Tyrosinase enzyme II.
a/ monophenolase
(monooxygenase) OH
function
OH
OH
+ O2 + AH2
+ A + H2O
R
R
b/ diphenolase
(oxidase)
function
OH
O
O
OH
2
+ O2
R
+ 2H2O
2
R
Blue copper oxidases I.
They catalyse the reduction of dioxygen to water by 4 electrons:
O2 + 4H+ + 4e− → 2 H2O
Structure:
In the resting state they contain min. 4 CuII:
I. + II. + 2 III.
The type II and III copper atoms usually forms a trimer unit.
Besides these, the proteins may contain further copper centres.
Important oxidases:
laccase (phenol oxidase): 4 copper atoms
ascorbic acid oxidase: 8 copper atoms
ceruloplasmin: 5 - 8 copper atoms (1 trimer + 2(5) type I)
Blue copper oxidases II.
The „trimer” activ centre of ascorbic acid oxidase:
The CuII → CuI reduction is accompanied by an increase in bond lengths.
The fourth Cu (type I) is situated rather far (1300 pm) from the trimer.
Superoxide dismutase (SOD) enzymes
They catalyse dysproportionation of the superoxide anion:
2 O2− → O2 + O22−
Main types:
CuZn-SOD: in eukaryots (cells with nucleus)
Fe-SOD: in prokaryots (cells without nucleus)
Mn-SOD: in prokaryots + mitochondrion
Ni-SOD: most recent (certain microorganisms)
Human SOD enzymes:
SOD 1: cytoplasm (CuZn)
SOD 2: mitochondrion (Mn)
SOD 3: extracellular (CuZn)
Characteristic features of CuZn-SOD:
Composition: 2 subunits (M 16.000/subunit)
(1Cu + 1Zn)/subunit
Structure:
Zn(II) (distorted tetrahedron), structure maker
Cu(II) (Type II: tetragonal), redoxy centre
All CuII complexes have low level of SOD activity.
Mechanism of CuZn-SOD
Zn(II): structure maker CoII, CdII or CuII may substitute (in vitro) it
without ceasing activity of the enzyme
Cu(II): participate actively in the redoxy process
without the metal ion the enzyme is not active
The catalytic reaction mechanism:
(temporary splitting of the Cu-His(61) bond)
Cu2+(His–)Zn2+ + O2– + H+
Cu+ + (HisH)Zn2+ + O2
Cu+ + O2– + H+ + (HisH)Zn 2+ Cu2+(His–)Zn2+ + H2O2
gross process:
2O2– + 2H+ H2O2 + O2
Structure of CuZn-SOD
–
2 O2 + 2
H+
H2O2 + O2
SOD
Cytochrome c oxidase
Function:
Terminal enzyme of the respiratory chain, catalysis the four
electron reduction of dioxygen to water.
Additionally, it generates a membrane proton gradient that
subsequently drives the synthesis of ATP.
Structure:
One of the most complex metalloproteins.
It consists of 13 subunits (M ~ 100.000),
some of them serve only to bind to the membrane.
The metal ion containing subunits:
Zn and Mg – structure makers
2 Fe: cytochrome a and cytochrome a3
3 Cu: CuA (2 copper) and CuB (1 copper)
Structure of CuA and CuB
CuA: mixed valence dimer
CuB: monomeric CuII centre, similar to a Type 2 copper, but
the His ligands are in trigonal pyramidal arrangement.
Peroxidases and the catalase
H2O2
Catalase or
peroxidase
H2O2
Compound I.
RH2
disproportion
ation
product
cit-cred, MnII, Cl-
{
cit-cox, MnIII, ClORH2
product
• In resting state haloperoxidases contain FeIII-hem,
• The peroxide oxidases the hem centre and an oxoferrilcentre
(FeIV=O) is formed,
• 1 electron comes from the hem or the protein and the radical
Compound I is formed,
• This reacts with H2O2 through disproportionation or RH2 substrate
or the redoxi partners (cytochrome, H2O2, Cl-, MnII) and reduced,
• The FeIV=O centre gives a water and returns to the FeIII-hem state.
Vanadium
Biological role
2. Vanadium containing enzymes
Haloperoxidases (Vilter, 1984)
isolated from red and brown algae species
Haloperoxidases
Chemical/Biological transformation of N-compounds
1. Nitrogen fixation (industrial):
metal oxide catalysts, T ~ 400 oC, p ~ 100-200 bar
N2 + 3 H2
2 NH3
2. Nitrogen fixation (biological):
(Mo containing nitrogenase enzyme)
N2 + 10H+ + 8 e− → 2 NH4+ + H2
3. Nitrification:
NH4+ + 2O2 → NO3− + H2O + 2H+
4. Denitrification:
(Mo containing nitrate reductase enzyme + Cu and hem)
2NO3− + 12H+ + 10e− → N2 + 6H2O
Nitrogenase and nitrogen fixation
1. Observation, Isolation:
Certain bacteria living in symbiosis with the root system of leguminous plants are able to utilise the dinitrogen of the air isolation of
Nitrogenase enzyme from these bacteria.
2. Model systems:
N2 complexes and their catalytic activity
e.g. RuII(NH3)5N22+,
CoI(N2)(H–)(PPh3)3
other metals and their oxidation state: (Mo0, W0, ReI, IrI, RhI.....)
activity: –
3. Structure and action of the nitrogenase:
N2 + 8H+ + 8e– + 16Mg-ATP 2NH3 + H2 + 16Mg-ADP + 16”P”
- Iron-molibden cofactor: FeMo-co
- Vanadium containing nitrogenase
- Metal free nitrogenases
Catalytic centres of the nitrogenase enzyme
It consists of two [Fe4S4] (P-cluster) and one [Fe4S4 – Fe3MoS3]
(M-cluster).
The N2 probably binds to the Mo, the energy of the reduction is
provided by the hydrolysis of ATP.
P-cluster
M-cluster
Vanadium-nitrogenase
(Hales and coworkers, 1986)
Azotobacter chroococcum isolated from
the A. vinelandii bacterium
It is active in the case of the lack of molibdenum
Xanthobacter autothrophycus accumulate
vanadium in Mo deficient environment.
Biological role
N2 fixation/reduction process
V-nitrogenase:
N2+10e+10H++ 24MgATP = 2NH3 + 3H2 + 24MgADP + 24Pi
+8H++ 16MgATP = 2NH + H + 16MgADP + 16P )
(NA2+8e
3
2 Fe -V-S k las zte r s ze rkie ze te
vanádium -nitroge náz e nzim be n található
Structure of Fe-V-S cluster in vanadium-nitrogenase enzyme
S
S
S
Fe
Fe
S
Fe
S
S
V
O
O
O
Tungsten containing enzymes
Tungsten is not considered as an essential element.
Tungsten containing enzymes were identified in some heatresistant organisms: these contains W-co (tungsten-cofactor)
Which corresponds to the Mo-co.
Its spreading in nature is uncertain, but is certainly not too frequent!
Based on their heatshock resistancy it can be assumed that they
appearred in the early stage of life but later the tungsten was
substituted by molibdenum.
(It might happened because of the difference in the availability
of the two metals or the kinetics of their substitution reactions).
Ellenőrző kérdések
1.
2.
3.
4.
5.
6.
Milyen kismolekulák aktiválására van szükség a
biológiai rendszerekben, és milyen fémionoknak van
ezen folyamatokban kitüntetett szerepe?
Hasonlítsa össze a dioxigenázokat és a
monooxigenázokat!
Hogyan alkalmazkodik a réz kémiai környezete a
biológiai funkcióhoz a réztartalmú fehérjékban? A
réztartalmú fehérjék fajtái és funkciói.
A vanádium biológiai szerepe.
A N2 fixálása.
A vas és a réztartalmú fehérjék funkcionális
összehasonlítása.