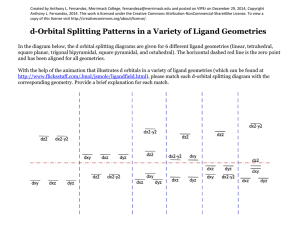
Slide 1
advertisement

Chemical Data and Computer-Aided Drug Discovery Mike Gilson School of Pharmacy mgilson@ucsd.edu 2-0622 Outline Overview of drug discovery Structure-based computational methods When we know the structure of the targeted protein Ligand-based computational methods When we don’t know the protein’s structure What is a drug? Small Molecule Drugs Aspirin Taxol Sildenafil (Viagra) Darunavir Glipizide (Glucotrol) Digoxin Nanoparticles (e.g., packaged small-molecule drugs) Doxil Abraxane (liposome package, extended circulation time,milder toxicity) (albumin-packaged taxol) http://www.doxil.com/about_doxil.html http://www.abraxane.com/professional/nab-technology.aspx Biopharmaceuticals Erythropoietin (EPO) Stabilized variant of a natural protein hormone http://www.ganfyd.org/index.php?title=Erythropoietin_beta Etanercept (Enbrel) Protein with TNF receptor + Ab Fc domain Scavenges TNF, diminishes inflammation http://en.wikipedia.org/wiki/File:Enbrel.jpg How are drugs discovered? Natural Products Aspirin Digoxin Taxol Pacific Yew Willow Foxglove How Aspirin Works Aspirin inflammation platelet activation platelet inactivation Biomolecular Pathways and Target Selection E.g. signaling pathways Target protein http://www.isys.uni-stuttgart.de/forschung/sysbio/insulin/index.html Empirical Path to Ligand Discovery Compound library (commercial, in-house, synthetic, natural) High throughput screening (HTS) Hit confirmation Lead compounds (e.g., µM Kd) Lead optimization (Medicinal chemistry) Animal and clinical evaluation Potent drug candidates (nM Kd) Compound Libraries Commercial (also in-house pharma) Academia Government (NIH) Computer-Aided Ligand Design Aims to reduce number of compounds synthesized and assayed Lower costs Less chemical waste Faster progress Scenario 1 Structure of Targeted Protein Known: Structure-Based Drug Discovery HIV Protease/KNI-272 complex Protein-Ligand Docking Structure-Based Ligand Design Docking software Potential function Search for structure of lowest energy Energy as function of structure VDW - + Screened Coulombic Dihedral Energy Determines Probability (Stability) Boltzmann distribution Probability Energy p( x) e x E ( x )/ RT Structure-Based Virtual Screening 3D structure of target Compound database (crystallography, NMR, modeling) Virtual screening (e.g., computational docking) Candidate ligands Ligand optimization Med chem, crystallography, modeling Experimental assay Ligands Drug candidates Fragmental Structure-Based Screening 3D structure of target “Fragment” library (crystallography, NMR, modeling) Fragment docking Compound design Experimental assay and ligand optimization Med chem, crystallography, modeling http://www.beilstein-institut.de/bozen2002/proceedings/Jhoti/jhoti.html Drug candidates Potential Functions for Structure-Based Design Energy as a function of structure Physics-Based Knowledge-Based Physics-Based Potentials Energy terms from physical theory Van der Waals interactions (shape fitting) Bonded interactions (shape and flexibility) Coulombic interactions (charge-charge complementarity) Hydrogen-bonding Common Simplifications Used in Physics-Based Docking Quantum effects approximated classically Protein often held rigid Configurational entropy neglected Influence of water treated crudely Proteins and Ligand are Flexible Protein Ligand Complex + DGo Binding Energy and Entropy 2e EBound / RT K EFree / RT 6e EFree Unbound states EBound Bound states DG RT ln K Ebound EFree RT ln 3 Energy part Entropy part Structure-Based Discovery Physics-oriented approaches Weaknesses Fully physical detail becomes computationally intractable Approximations are unavoidable Parameterization still required Strengths Interpretable, provides guides to design Broadly applicable, in principle at least Clear pathways to improving accuracy Status Useful, far from perfect Multiple groups working on fewer, better approxs Force fields, quantum Flexibility, entropy Water effects Moore’s law: hardware improving Knowledge-Based Docking Potentials Ligand carboxylate Aromatic stacking Probability Energy Boltzmann: p(r ) e E ( r )/ RT Inverse Boltzmann: E(r ) RT ln p(r ) Example: ligand carboxylate O to protein histidine N 1. 2. 3. 4. Find all protein-ligand structures in the PDB with a ligand carboxylate O For each structure, histogram the distances from O to every histidine N Sum the histograms over all structures to obtain p(rO-N) Compute E(rO-N) from p(rO-N) Knowledge-Based Docking Potentials “PMF”, Muegge & Martin, J. Med. Chem. 42:791, 1999 A few types of atom pairs, out of several hundred total Nitrogen+/Oxygen- Aromatic carbons Aliphatic carbons Atom-atom distance (Angstroms) Eprot lig Evdw pairs (ij ) Etype(ij ) (rij ) Structure-Based Discovery Knowledge-based potentials Weaknesses Accuracy limited by availability of data Accuracy may also be limited by overall approach Strengths Relatively easy to implement Computationally fast Status Useful, far from perfect May be at point of diminishing returns Limitations of Knowledge-Based Potentials 1. Statistical limitations (e.g., to pairwise potentials) 100 bins for a histogram of O-N & O-C distances 10 bins for a histogram of O-N distances r1 r2 … r10 rO-C rO-N rO-N 2. Even if we had infinite statistics, would the results be accurate? (Is inverse Boltzmann quite right? Where is entropy?) Scenario 2 Structure of Targeted Protein Unknown: Ligand-Based Drug Discovery e.g. MAP Kinase Inhibitors Using knowledge of existing inhibitors to discover more Why Look for Another Ligand if You Already Have Some? Experimental screening generated some ligands, but they don’t bind tightly A company wants to work around another company’s chemical patents An high-affinyt ligand is toxic, is not well-absorbed, etc. Ligand-Based Virtual Screening Compound Library Known Ligands Molecular similarity Machine-learning Etc. Candidate ligands Optimization Med chem, crystallography, modeling Assay Actives Potent drug candidates Sources of Data on Known Ligand Journals, e.g., J. Med. Chem. Some Binding and Chemical Activity Databases PubChem (NIH) pubchem.ncbi.nlm.nih.gov ChEMBL (EMBL) www.ebi.ac.uk/chembl BindingDB (UCSD) www.bindingdb.org BindingDB www.bindingdb.org Finding Protein-Ligand Data in BindingDB e.g., by Name of Protein “Target” e.g., by Ligand Draw Search Sample Query Results BindingDB to PDB PDB to BindingDB Download data in machine-readable format Sample Query Results Machine-Readable Chemical Format Structure-Data File (SDF) SDF Format Defines Chemical Bonds PDB Format Lacks Chemical Bonding There are Many Other Chemical File Formats Interconvert with Babel Chemical Similarity Ligand-Based Drug-Discovery Compounds (available/synthesizable) Similar Test experimentally Don’t bother Chemical Fingerprints Binary Structure Keys … Molecule 1 Molecule 2 Chemical Similarity from Fingerprints Tanimoto Similarity or Jaccard Index, T NI T 0.25 NU Intersection NI=2 Union NU=8 Molecule 1 Molecule 2 Hashed Chemical Fingerprints Based upon paths in the chemical graph 1-atom paths: C F N H S O 2-atom paths: F-C C-C C-N C-S S-O C-H 3-atom paths: F-C-C C-C-N C-N-H C-S-O Each path sets a pseudo-random bit-pattern in a very long molecular fingerprint C S-O etc. Maximum Common Substructure Ncommon=34 Potential Drawbacks of Plain Chemical Similarity May miss good ligands by being overly conservative Too much weight on irrelevant details Scaffold Hopping Identification of synthetic statins by scaffold hopping Zhao, Drug Discovery Today 12:149, 2007 Abstraction and Identification of Relevant Compound Features Ligand shape Pharmacophore models Chemical descriptors Statistics and machine learning Pharmacophore Models Φάρμακο (drug) + Φορά (carry) A 3-point pharmacophore Bulky hydrophobe 3.2 ±0.4 Å +1 Aromatic Molecular Descriptors More abstract than chemical fingerprints Physical descriptors molecular weight charge dipole moment number of H-bond donors/acceptors number of rotatable bonds hydrophobicity (log P and clogP) Topological branching index measures of linearity vs interconnectedness Etc. etc. Rotatable bonds A High-Dimensional “Chemical Space” Each compound is at a point in an n-dimensional space Descriptor 3 Compounds with similar properties are near each other Descriptor 2 Point representing a compound in descriptor space Statistics and Machine Learning Some examples Partial least squares Support vector machines Genetic algorithms for descriptor-selection Summary Overview of drug discovery Computer-aided methods Structure-based Ligand-based Interaction potentials Physics-based Knowledge-based (data driven) Ligand-protein databases, machine-readable chemical formats Ligand similarity and beyond Mike Gilson, School of Pharmacy, mgilson@ucsd.edu, 2-0622 Activities and Discussion Topics BindingDB: Advil Machine-readable format, Binding activities PDB/BindingDB 2ONY at PDB BindingDB Substructure search Related data Similarity search Combined computational approaches (physics + knowledge)-based docking potentials (ligand + structure)-based computational discovery Other data-driven methods where it may be hard to get enough statistics Validation of computational methods Protein-ligand databases: getting data and assessing data quality Drug Discovery Pipeline (One Model) Target identification Target validation Assay development Phase I Clinical (safety, metab, PK) Lead compound (ligand) discovery Phase II Clinical (efficacy) Lead optimization Phase III Clinical (comparison with existing therapy) Animal Pharmacokinetics, Toxicity Updated Knowledge-Based PMF Potential Muegge J. Med. Chem. 49: 5895, 2006