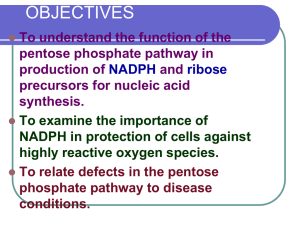
absolute quantification
advertisement

Proteored Multicentric Experiment 8 (PME8) Quantitative Targeted Analysis in Proteomics. An Assesment Study (QTAPAS) ProteoRed WG1-WG2 Meeting Pamplona, December 10th 2013 Total 27 Participant Laboratories (February- April 2013) MS Instrument Flow rate LC Column Gradient time Scheduled VHIO X 4000QTrap 300nL/min 75 µm x 15 cm 90 no 2 IACS X 4000QTrap 300 nL/min 75 µm x 15 cm 90 no 3 5500QTrap 15 µl/min 500 µm x 10 cm 20 yes # Lab SRM 1 MPM ABSciex X 4 INIBIC X 5500QTrap 300 nL/min 75 µm x 15 cm 100 no 5 UPF-CRG X 5500QTrap 300 nL/min 75 µm x 12 cm 35 yes 6 CIMA X 5500QTrap 300 nL/min 75 µm x 15 cm 90 no 7 CNB X 5500Qtrap 300 nL/min 75 µm x 15 cm 90 no 8 UCM-PCM X 5500QTrap 300 nL/min 75 µm x 15 cm 90 no 9 CSPI-Lund X TSQ Vantage 300 nL/min 75 µm x 12 cm 60 yes X TSQ Vantage 300 nL/min 75 µm x 15 cm 30 yes X TSQ Vantage 300 nL/min 75 µm x 15 cm 60 yes 10 11 Groningen 12 X TSQ Vantage 300 nL/min yes UV X 5600TripleTOF 300 nL/min 75 µm x 15 cm 75 µm x 12.3 cm 90 13 90 no 14 Navarra Bio. X 5500QTrap 300 nL/min 75 µm x 15 cm 90 no 15 Sigma X 5500QTrap 4 µL/min 500 µm x 15 cm 45 no 16 UA X Agilent 6490 300 nL/min 75 µm x 15 cm 90 yes 17 Turku X TSQVantage 300 nL/min 75 µm x 15 cm 90 yes X 4000 QTrap 300 nL/min 75 µm x 15 cm 90 no X Agilent 6490 300 nL/min 75 µm x 15 cm 90 no X 5500QTrap 250 µL/min 75 µm x 15 cm 90 no 18 19 UVIC 20 UiB 21 EHU X Q-Exactive 300 nL/min 50 µm x 15 cm 90 22 IRBB X Synapt G1 250 nL/min 75 µm x 25 cm 90 23 CIB X Orbitrap Velos 250 nL/min 75 µm x 15 cm 90 24 X Orbitrap Velos 300 nL/min 75 µm x 10 cm 90 25 CBMSO CICSalamanca X Orbitrap Velos 400 nL/min 75 µm x 10 cm 90 26 LPCSIC-UAB X Orbitrap Velos 0.8 µL/min 100 µm x 15 cm 90 27 CCiT-UB X Orbitrap Velos 400 nL/min 75 µm x 10 cm 90 28 PCB X Orbitrap Velos 250 µL/min 75 µm x 25 cm 90 29 CMU-Geneva X Orbitrap Velos 250 µL/min 75 µm x 25 cm 90 30 CICbioGUNE X Orbitrap XL ETD 300 nL/min 75 µm x 20 cm 90 20 SRM data sets 10 PRM data sets: 3 QTOF 7 OT TQ1 TQ2 TQ3 TQ4 TQ5 TQ6 TQ7 TQ8 TQ9 TQ10 TQ11 TQ12 TQ13 TQ14 TQ15 TQ16 TQ17 TQ18 TQ19 TQ20 QTOF21 QTOF22 QTOF23 OT24 OT25 OT26 OT27 OT28 OT29 OT30 VHIO IACS UVIC1 (Vancouver) AbSciex INIBIC UPF-CRG CIMA CNB UCM-PCM SIGMA UiB(Bergen) NavarraBiomed UA UVIC2 (Vancouver) Turku CSPI (Lund) Groningen 30 Groningen 60 Groningen 90 Waters UV EHU IRB CIB CBMSO CIC-Salamanca CCiT PCB LPCSIC-UAB CiCBiogune MRM x x x x x x x x x x x x x x x x x x x x PRM x x x x x x x x x x Mass Spectrometer 4000QTrap 4000QTrap 4000QTrap 5500QTrap 5500QTrap 5500QTrap 5500QTrap 5500Qtrap 5500QTrap 5500QTrap 5500QTrap 5500QTrap Agilent 6490 QQQ Agilent 6490 QQQ TSQ Vantage TSQ Vantage TSQ Vantage TSQ Vantage TSQ Vantage Xevo TQ-S 5600TripleTOF Q-Exactive Waters Synapt G1 LTQ Orbitrap Velos LTQ Orbitrap Velos LTQ Orbitrap Velos LTQ Orbitrap Velos LTQ Orbitrap Velos LTQ Orbitrap Velos Orbitrap XL ETD TQ QTOF OT SIGMA-ALDRICH MSQC1 - 6 Digested proteins : 25-fold concentration range – 3 concentration tiers Quantified UV before digestion - 2-3 labeled peptides/ protein L/H ratios 0.2-50 Quantified AAA SAMPLE SET Yeast Digest + Spiked MSQC1, 5 different concentrations Tier Protein CAH1 1 CAH2 NADPH 2 CRP PPIA 3 CATA fmol / mg yeast digest sample A sample B sample C sample D sample E 20 40 100 200 1000 20 40 100 200 1000 4 8 20 40 200 4 8 20 40 200 0.8 1.6 4 8 40 0.8 1.6 4 8 40 fmol / microgram Yeast Lysate Digest LABELED PEPTIDES Tier 1 2 3 ratio L/H CAH1_GGPFSDSY[R] 1 Labeled AQUA Peptides fmol/m g Yeast digest CAH1_VLDALQAI[K] 2 CAH2_AVQQPDGLAVLGIFL[K] 10 A B C CAH2_SADFTNFDP[R] 50 NADPH_EGHLSPDIVAEQ[K] 1 GGPFSDSY[R] 20 40 CAH1 VLDALQAI[K] 10 20 NADPH_ALIVLAHSE[R] 2 AVQQPDGLAVLGIFL[K] 2 4 CRP_ESDTSYVSL[K] 10 CAH2 SADFTNFDP[R] 0.4 0.8 CRP_GYSIFSYAT[K] 50 EGHLSPDIVAEQ[K] 0.5 4 8 PPIA_FEDENFIL[K] NADPH ALIVLAHSE[R] 2 4 PPIA_VSFELFAD[K] 1 ESDTSYVSL[K] 0.4 0.8 PPIA_TAENF[R] 2 CRP GYSIFSYAT[K] 0.08 0.16 CATA_GAGAFGYFEVTHDIT[K] 0.2 FEDENFIL[K] 1.6 3.2 CATA_FSTVAGESGSADTV[R] 10 PPIA VSFELFAD[K] 0.8 1.6 CATA_NLSVEDAA[R] 50 TAENF[R] 0.4 0.8 GAGAFGYFEVTHDIT[K] 4 8 CATA FSTVAGESGSADTV[R] 0.08 0.16 NLSVEDAA[R] 0.016 0.032 Peptide D 100 50 10 2 20 10 2 0.4 8 4 2 20 0.4 0.08 E 200 1000 100 500 20 100 4 20 40 200 20 100 4 20 0.8 4 16 80 8 40 4 20 40 200 0.8 4 0.16 0.8 QTAPAS - PME8 SRM ANALYSIS RECOMMENDED GUIDELINES 1- LC conditions: -Nano LC- system: Column: 75 mm x 15 cm C18 Sample load: 1 mg yeast digest/injection Flow rate: 300-500 nL /min Gradient: 0-35% Acetonitrile in 90 min 3- Analysis design: - Samples A through E Replica 1 (from most diluted to most concentrated) - Blank runs (enough to secure clean baseline) - Samples A through E Replica 2 - Blank runs - Samples A through E Replica 3 http://www.proteored.org/PME8_main.asp LINEARITY OF RESPONSE Average of 30 datasets – Error bars: Std Dev. Tier 1 CAH1 CAH1 10000 1200 y = 1.0153x R² = 1 800 1000 y = 0.9987x R² = 1 600 400 log measured fmol measured fmol CAH1_GGPFSDSYR CAH1_VLDALQAIK CAH1_GGPFSDSYR CAH1_VLDALQAIK 1000 100 10 200 1 0 0 200 400 600 theoretical fmol 800 1000 1 10 100 log theoretical fmol 1000 10000 Tier 2 NADPH NADPH 1000 200 NADPH_EGHLSPDIVAEQK NADPH_ALIVLAHSER y = 0.8673x R² = 0.9998 120 80 y = 0.4689x R² = 0.9999 40 log measured fmol measured fmol 160 NADPH_EGHLSPDIVAEQK NADPH_ALIVLAHSER 100 10 1 0 0 40 80 120 theoretical fmol 160 200 1 10 100 log theoretical fmol 1000 Tier 3 PPIA PPIA 30 100 25 y = 0.5552x R² = 0.9999 20 15 log measured fmol measured fmol PPIA_FEDENFILK PPIA_VSFELFADK PPIA_FEDENFILK PPIA_VSFELFADK y = 0.5579x R² = 0.9993 10 10 1 0.1 1 10 5 0 0 0 5 10 15 20 25 30 theoretical fmol 35 40 45 log theoretical fmol 100 ABSOLUTE QUANTIFICATION INTER LABORATORY VARIABILITY %CV 30 datasets Average of 30 datasets , three replicas averaged Sample A Peptide theor. fmol ALL MEASUREMENTS B C D E A B Average fmol measured C D E CV(%) 20 40 100 200 1000 CAH1_GGPFSDSYR 20.41 40.65 102.30 202.87 1015.23 4.80 6.29 4.81 4.90 4.82 CAH1_VLDALQAIK 20.58 41.02 96.75 201.47 998.66 13.19 6.02 12.89 6.93 10.74 CAH2_AVQQPDGLAVLGIFLK 16.93 33.76 87.94 154.83 852.65 45.22 28.31 27.13 19.93 10.87 CAH2_SADFTNFDPR 19.58 41.44 107.41 221.13 1125.46 19.80 19.19 18.15 10.02 12.65 4.00 8.00 20.00 40.00 200.00 NADPH_EGHLSPDIVAEQK theor. fmol 3.66 7.15 19.41 35.38 173.10 27.02 20.71 20.90 13.50 11.60 NADPH_ALIVLAHSER 1.85 3.76 9.25 19.35 93.68 22.78 15.78 15.45 11.50 7.86 CRP_ESDTSYVSLK 3.92 7.87 18.43 36.81 185.79 22.05 28.90 9.89 6.88 7.42 CRP_GYSIFSYATK 2.16 4.47 13.32 28.76 149.52 47.30 27.58 13.06 14.66 9.00 0.80 1.60 4.00 8.00 40.00 PPIA_FEDENFILK 0.50 0.93 2.23 4.33 22.23 18.92 18.67 22.08 12.87 15.91 PPIA_VSFELFADK 0.72 1.02 2.59 4.48 22.27 48.45 37.92 28.81 14.34 14.65 CATA_GAGAFGYFEVTHDITK 1.15 0.53 1.47 2.56 8.29 75.82 48.13 67.03 45.56 24.25 CATA_FSTVAGESGSADTVR 0.55 0.92 2.49 4.69 22.13 113.34 41.87 33.56 18.53 11.48 CATA_NLSVEDAAR 0.27 0.63 2.33 4.96 27.22 79.90 61.85 41.77 30.83 12.96 theor. fmol ABSOLUTE QUANTIFICATION INTER LABORATORY VARIABILITY Sample A Peptide theor. fmol A B C D E CV(%) 20 40 100 200 1000 20.50 40.96 102.29 204.21 1024.68 5.42 5.31 5.26 5.33 5.38 CAH1_VLDALQAIK 21.01 41.29 99.34 205.31 1004.96 13.47 5.82 12.17 4.68 9.64 CAH2_AVQQPDGLAVLGIFLK 15.71 33.27 89.06 168.98 844.86 40.58 24.68 25.32 9.58 9.14 CAH2_SADFTNFDPR 20.69 44.12 114.38 228.20 1154.97 11.26 13.75 9.99 4.11 4.57 4.00 8.00 20.00 40.00 200.00 NADPH_EGHLSPDIVAEQK 3.80 7.38 19.10 35.85 173.37 24.82 19.31 15.22 12.91 12.23 NADPH_ALIVLAHSER 2.07 3.89 9.55 19.59 95.10 13.57 16.00 15.81 12.06 6.00 CRP_ESDTSYVSLK 3.83 7.40 18.79 37.06 187.42 17.24 10.46 6.62 5.15 6.82 CRP_GYSIFSYATK 2.17 4.38 13.17 29.62 149.62 52.60 30.63 13.75 13.10 9.50 0.80 1.60 4.00 8.00 40.00 PPIA_FEDENFILK 0.51 0.90 2.15 4.29 21.32 18.58 16.83 13.64 13.98 9.71 PPIA_VSFELFADK 0.72 1.06 2.38 4.47 22.30 41.29 31.94 19.19 15.42 15.93 CATA_GAGAFGYFEVTHDITK 1.12 0.47 1.24 2.25 8.04 80.74 46.49 79.39 46.25 25.30 CATA_FSTVAGESGSADTVR 0.62 0.90 2.27 4.79 22.02 113.39 18.95 23.42 17.60 12.70 CATA_NLSVEDAAR 0.30 0.69 2.24 4.95 27.12 76.51 59.91 44.70 33.85 8.64 A B A B C D theor. fmol Sample Peptide theor. fmol ORBITRAP C D E Average fmol measured E CV(%) 20 40 100 200 1000 CAH1_GGPFSDSYR 20.01 39.82 101.79 198.71 1001.52 1.84 10.81 4.09 3.48 1.37 CAH1_VLDALQAIK 18.22 39.73 86.40 181.99 986.23 11.41 9.08 11.49 10.75 11.83 CAH2_AVQQPDGLAVLGIFLK 19.66 29.31 73.73 124.25 934.06 65.94 29.58 17.19 29.67 17.50 CAH2_SADFTNFDPR 15.13 29.72 90.46 186.49 1002.49 37.49 18.12 29.96 24.88 27.64 4.00 8.00 20.00 40.00 200.00 NADPH_EGHLSPDIVAEQK 3.06 5.79 22.60 27.55 161.07 10.04 21.75 53.26 NADPH_ALIVLAHSER 1.33 3.29 8.59 18.44 88.68 17.52 7.65 13.20 10.16 17.06 CRP_ESDTSYVSLK 3.69 7.49 15.61 32.95 167.97 9.04 49.12 8.07 17.33 9.53 CRP_GYSIFSYATK 1.97 4.54 14.65 25.03 theor. fmol N=7 E Average fmol measured CAH1_GGPFSDSYR theor. fmol N=20 TRIPLE-QUADRUPOLE C D B theor. fmol 15.11 0.80 1.60 4.00 8.00 40.00 PPIA_FEDENFILK 0.41 0.96 2.05 4.48 24.72 6.74 18.29 10.26 5.88 24.20 PPIA_VSFELFADK 0.83 0.97 2.96 4.66 22.04 70.22 57.51 41.07 14.31 14.98 CATA_GAGAFGYFEVTHDITK 0.79 0.45 1.56 2.40 9.19 56.87 14.18 73.57 29.09 21.71 CATA_FSTVAGESGSADTVR 0.37 0.80 2.98 3.94 22.38 12.72 9.71 41.03 25.04 8.84 CATA_NLSVEDAAR 0.14 0.40 2.91 5.30 24.86 109.32 71.70 32.27 10.70 21.96 ABSOLUTE QUANTIFICATION INTER LABORATORY VARIABILITY Sample A Peptide theor. fmol N=20 E A B Average fmol measured C D E CV(%) 20 40 100 200 1000 CAH1_GGPFSDSYR 20.50 40.96 102.29 204.21 1024.68 5.42 5.31 5.26 5.33 5.38 CAH1_VLDALQAIK 21.01 41.29 99.34 205.31 1004.96 13.47 5.82 12.17 4.68 9.64 CAH2_AVQQPDGLAVLGIFLK 15.71 33.27 89.06 168.98 844.86 40.58 24.68 25.32 9.58 9.14 CAH2_SADFTNFDPR 20.69 44.12 114.38 228.20 1154.97 11.26 13.75 9.99 4.11 4.57 4.00 8.00 20.00 40.00 200.00 NADPH_EGHLSPDIVAEQK 3.80 7.38 19.10 35.85 173.37 24.82 19.31 15.22 12.91 12.23 NADPH_ALIVLAHSER 2.07 3.89 9.55 19.59 95.10 13.57 16.00 15.81 12.06 6.00 CRP_ESDTSYVSLK 3.83 7.40 18.79 37.06 187.42 17.24 10.46 6.62 5.15 6.82 CRP_GYSIFSYATK 2.17 4.38 13.17 29.62 149.62 52.60 30.63 13.75 13.10 9.50 0.80 1.60 4.00 8.00 40.00 PPIA_FEDENFILK 0.51 0.90 2.15 4.29 21.32 18.58 16.83 13.64 13.98 9.71 PPIA_VSFELFADK 0.72 1.06 2.38 4.47 22.30 41.29 31.94 19.19 15.42 15.93 CATA_GAGAFGYFEVTHDITK 1.12 0.47 1.24 2.25 8.04 80.74 46.49 79.39 46.25 25.30 CATA_FSTVAGESGSADTVR 0.62 0.90 2.27 4.79 22.02 113.39 18.95 23.42 17.60 12.70 CATA_NLSVEDAAR 0.30 0.69 2.24 4.95 27.12 76.51 59.91 44.70 33.85 8.64 Sample A B C D E theor. fmol theor. fmol Peptide theor. fmol Q-TOF C D E A B Average fmol measured CV(%) 20 40 100 200 1000 CAH1_GGPFSDSYR 20.44 40.20 102.97 200.38 981.40 3.27 3.00 3.97 3.47 2.71 CAH1_VLDALQAIK 21.19 41.07 95.07 204.44 977.43 3.65 1.80 14.84 4.31 19.57 CAH2_AVQQPDGLAVLGIFLK 19.40 43.28 107.15 139.73 792.83 7.23 30.28 32.62 27.68 4.19 CAH2_SADFTNFDPR 20.71 43.44 90.52 212.91 1182.04 21.38 9.12 31.40 0.71 11.31 58.66 23.84 16.67 13.41 6.86 theor. fmol N=3 TRIPLE-QUADRUPOLE C D B 4.00 8.00 20.00 40.00 200.00 NQO1_EGHLSPDIVAEQK 3.63 7.54 18.53 35.77 179.64 NQO1_ALIVLAHSER 1.68 3.56 8.28 18.56 89.12 CRP_ESDTSYVSLK 4.86 11.05 19.15 37.98 188.45 48.40 37.45 CRP_GYSIFSYATK 2.13 4.96 14.12 24.56 148.97 16.25 18.92 0.80 1.60 4.00 8.00 40.00 PPIA_FEDENFILK 0.51 1.14 3.16 4.65 24.84 PPIA_VSFELFADK 0.42 0.94 3.32 4.26 22.47 CATA_GAGAFGYFEVTHDITK 2.70 1.10 2.25 5.01 CATA_FSTVAGESGSADTVR 0.28 1.16 3.54 5.02 22.84 CATA_NLSVEDAAR 0.28 0.75 1.81 4.39 31.48 theor. fmol 3.04 24.77 15.39 3.18 5.60 7.70 21.92 7.51 39.59 25.76 7.97 97.18 93.92 49.89 14.23 0.20 20.08 fmol measured TQ Sample OT QTOF D E D E D E 200 1000 200 1000 200 1000 CAH1_GGPFSDSYR 204.21 1024.68 198.71 1001.52 200.38 981.40 CAH1_VLDALQAIK 205.31 1004.96 181.99 986.23 204.44 977.43 CAH2_AVQQPDGLAVLGIFLK 168.98 844.86 124.25 934.06 139.73 792.83 CAH2_SADFTNFDPR 228.20 1154.97 186.49 1002.49 212.91 1182.04 theor. fmol 40.00 200.00 40.00 200.00 40.00 200.00 NADPH_EGHLSPDIVAEQK 35.85 173.37 27.55 161.07 35.77 179.64 NADPH_ALIVLAHSER 19.59 95.10 18.44 88.68 18.56 89.12 CRP_ESDTSYVSLK 37.06 187.42 32.95 167.97 37.98 188.45 CRP_GYSIFSYATK 29.62 149.62 25.03 24.56 148.97 8.00 40.00 8.00 40.00 8.00 40.00 PPIA_FEDENFILK 4.29 21.32 4.48 24.72 4.65 24.84 PPIA_VSFELFADK 4.47 22.30 4.66 22.04 4.26 22.47 CATA_GAGAFGYFEVTHDITK 2.25 8.04 2.40 9.19 5.01 CATA_FSTVAGESGSADTVR 4.79 22.02 3.94 22.38 5.02 22.84 CATA_NLSVEDAAR 4.95 27.12 5.30 24.86 4.39 31.48 Peptide theor. fmol theor. fmol %CV TQ Sample OT QTOF D E D E D E 200 1000 200 1000 200 1000 CAH1_GGPFSDSYR 5.33 5.38 3.48 1.37 3.47 2.71 CAH1_VLDALQAIK 4.68 9.64 10.75 11.83 4.31 19.57 CAH2_AVQQPDGLAVLGIFLK 9.58 9.14 29.67 17.50 27.68 4.19 CAH2_SADFTNFDPR 4.11 4.57 24.88 27.64 0.71 11.31 40.00 200.00 40.00 200.00 40.00 200.00 NADPH_EGHLSPDIVAEQK 12.91 12.23 15.11 13.41 6.86 NADPH_ALIVLAHSER 12.06 6.00 10.16 17.06 CRP_ESDTSYVSLK 5.15 6.82 17.33 9.53 CRP_GYSIFSYATK 13.10 9.50 8.00 40.00 8.00 40.00 PPIA_FEDENFILK 13.98 9.71 5.88 24.20 25.76 PPIA_VSFELFADK 15.42 15.93 14.31 14.98 7.97 CATA_GAGAFGYFEVTHDITK 46.25 25.30 29.09 21.71 CATA_FSTVAGESGSADTVR 17.60 12.70 25.04 8.84 CATA_NLSVEDAAR 33.85 8.64 10.70 21.96 Peptide theor. fmol theor. fmol theor. fmol 3.18 5.60 7.70 21.92 7.51 8.00 40.00 14.23 0.20 20.08 ABSOLUTE QUANTIFICATION Comparison by type of measurement Sample E (1000 fmol) 200 950 25 24 fmol measured 1100 1000 CATA_FSTVAGESGSADTVR NADPH_EGHLSPDIVAEQK 220 fmol measured fmol measured CAH1_GGPFSDSYR 1150 1050 Sample E (40 fmol) Sample E (200 fmol) 180 160 140 900 OT QTOF 21 19 TQ Sample A (20 fmol) 22 20 120 TQ 23 OT TQ QTOF CAH1_GGPFSDSYR CATA_FSTVAGESGSADTVR NADPH_EGHLSPDIVAEQK 7 22 6 QTOF Sample A (0.8 fmol) Sample A (4 fmol) 23 OT 0.7 0.6 20 fmol measured fmol measured fmol measured 0.5 21 5 4 0.4 0.3 0.2 19 3 0.1 18 0 2 TQ OT QTOF TQ OT QTOF TQ OT QTOF ABSOLUTE QUANTIFICATION INTRA- vs INTER LABORATORY VARIABILITY Average, %CV 3 replicas - Average, %CV 30 datasets 30 Sample E 25 %CV 20 15 10 Inter-Lab %CV Intra-Lab %CV 5 0 INTRA- vs INTER LABORATORY VARIABILITY Average, %CV 3 replicas 120 - Average, %CV 30 datasets Sample A 100 %CV 80 60 40 Inter-Lab %CV Intra-Lab %CV 20 0 REPRODUCIBILITY OF TRANSITION PATTERN Transition signal distribution Average 3 replicas 1.2 CAH1_VLDALQAIK 1 Normalized dot product to average distribution 1.02 CAH1_VLDALQAI[K] 1 0.8 TQ B 0.98 0.6 QTOF B OT B 0.96 TQ E 0.4 0.94 QTOF E 0.2 OT E 0.92 0 0.9 1.2 1 0.8 CATA_FSTVAGESGSADTVR CATA_FSTVAGESGSADTVR 1 0.95 TQ B QTOF B 0.6 OT B 0.9 0.4 0.2 0 TQ E QTOF E 0.85 0.8 OT E REPRODUCIBILITY OF TRANSITION PATTERN 1.2 CAH2_AVQQPDGLAVLGIFLK 1 1 0.9 0.8 0.8 0.6 0.4 0.2 0 CAH2_AVQQPDGLAVLGIFL[K] 0.7 TQ E 0.6 QTOF E 0.5 OT E 0.4 0.3 0.2 INTRA- vs INTER LABORATORY VARIABILITY 120 Sample A 100 %CV 80 60 40 Inter-Lab %CV Intra-Lab %CV 20 0 Analysis of pure MSQC1 (without yeast background) For comparison: Possible effects of complex background 3 Vials x 10 mg 200 ng injection – 80 fmol min Proteored Multicentric Experiment 8: Quantitative Targeted Analysis in Proteomics. An Assessment Study. (PME8-QTAPAS) Francesc Canals1 , Joan-Josep Bech-Serra1, Núria Colomé-Calls1, Salvador Martínez-Bartolomé2, Jim J. Walters3, Kevin B. Ray3, Juan-Pablo Albar2, ProteoRed-ISCIII Consortium4 1 Proteomics Laboratory, Vall d’Hebron Institute of Oncology (VHIO) Vall d’Hebron University Hospital, 08035 Barcelona, Spain Centro Nacional de Biotecnologia-CSIC, Madrid, Spain Sigma-Aldrich, St. Louis, MO 4 ProteoRed-ISCIII Consortium. Spanish Networked Proteomics Platform fcanals@vhio.net ; jpalbar@proteored.org 2 3 SAMPLE SET STUDY DESIGN AND OBJECTIVE Analysis conditions - 3 replicate nLC-MS runs, 90 min gradient , batchwise (3 x [A-E ]) - SRM: Monitoring (14 +14) peptides x 3 transitions = 72 transitions - MPM: Monitoring (14+14) peptides, 30 precursor ions Participant Laboratories fmol / mg yeast digest sample A sample B sample C sample D sample E 20 40 100 200 1000 20 40 100 200 1000 4 8 20 40 200 4 8 20 40 200 0.8 1.6 4 8 40 0.8 1.6 4 8 40 Tier Protein CAH1 1 CAH2 NADPH 2 CRP PPIA 3 CATA Tier Peptide CAH1_GGPFSDSY[R] CAH1_VLDALQAI[K] 1 CAH2_AVQQPDGLAVLGIFL[K] CAH2_SADFTNFDP[R] NADPH_EGHLSPDIVAEQ[K] NADPH_ALIVLAHSE[R] 2 CRP_ESDTSYVSL[K] CRP_GYSIFSYAT[K] PPIA_FEDENFIL[K] PPIA_VSFELFAD[K] • Set of 5 different samples A-E, prepared by spiking different amounts of the Sigma-Aldrich MSQC1 standard into a yeast lysate digest. • The samples contain tryptic digests of 6 human proteins, distributed in three concentration tiers, as shown in the table. Amounts indicated as fmol spiked protein/ microgram of yeast lysate • Additionally, the samples contain isotopically labeled peptides for each of the human proteins, in different ratios to the corresponding unlabeled peptides, ranging form 1:0.2 to 1:50, as indicated in the table. PPIA_TAENF[R] 3 CATA_GAGAFGYFEVTHDIT[K] CATA_FSTVAGESGSADTV[R] CATA_NLSVEDAA[R] ratio L/H 1 2 10 50 1 2 10 50 0.5 1 2 0.2 10 50 • 20 SRM Triple Quadrupole Data Sets • 3 Q-TOF + 7 ORBITRAP MPM Data Sets INTRA- LABORATORY VARIABILITY RESULTS. ABSOLUTE QUANTIFICATION. INTER- LABORATORY/MEASUREMENT TYPE VARIABILITY A B TRIPLE-QUADRUPOLE C D E A B 5.42 5.31 C Average fmol measured D E 5.33 5.38 Sample CV(%) 20 40 100 200 1000 20.50 40.96 102.29 204.21 1024.68 A B Peptide 30 ALL MEASUREMENTS C D E A B 4.80 6.29 C Average fmol measured theor. fmol 5.26 CAH1_GGPFSDSYR Sample E D E 4.90 4.82 CV(%) 20 40 100 200 1000 20.41 40.65 102.30 202.87 1015.23 25 4.81 CAH1_VLDALQAIK 21.01 41.29 99.34 205.31 1004.96 13.47 5.82 12.17 4.68 9.64 CAH1_VLDALQAIK 20.58 41.02 96.75 201.47 998.66 13.19 6.02 12.89 6.93 10.74 CAH2_AVQQPDGLAVLGIFLK 15.71 33.27 89.06 168.98 844.86 40.58 24.68 25.32 9.58 9.14 CAH2_AVQQPDGLAVLGIFLK 16.93 33.76 87.94 154.83 852.65 45.22 28.31 27.13 19.93 10.87 CAH2_SADFTNFDPR 20.69 44.12 114.38 228.20 1154.97 11.26 13.75 9.99 4.11 4.57 CAH2_SADFTNFDPR 19.58 41.44 107.41 221.13 1125.46 19.80 19.19 18.15 10.02 12.65 24.82 12.23 NADPH_EGHLSPDIVAEQK 27.02 11.60 theor. fmol NADPH_EGHLSPDIVAEQK 4.00 8.00 20.00 40.00 200.00 3.80 7.38 19.10 35.85 173.37 15.22 12.91 9.55 19.59 95.10 13.57 16.00 15.81 12.06 18.79 37.06 187.42 17.24 10.46 6.62 5.15 4.38 13.17 29.62 149.62 52.60 30.63 13.75 13.10 1.60 4.00 8.00 40.00 2.07 3.83 CRP_GYSIFSYATK theor. fmol theor. fmol 19.31 3.89 7.40 2.17 0.80 NADPH_ALIVLAHSER CRP_ESDTSYVSLK 4.00 8.00 20.00 40.00 200.00 3.66 7.15 19.41 35.38 173.10 20.71 20.90 13.50 3.76 9.25 19.35 93.68 22.78 15.78 15.45 11.50 7.87 18.43 36.81 185.79 22.05 28.90 9.89 6.88 2.16 4.47 13.32 28.76 149.52 47.30 27.58 13.06 14.66 0.80 1.60 4.00 8.00 40.00 6.00 NADPH_ALIVLAHSER 1.85 6.82 CRP_ESDTSYVSLK 3.92 9.50 CRP_GYSIFSYATK theor. fmol 7.86 0.90 2.15 4.29 21.32 18.58 16.83 13.64 13.98 9.71 PPIA_FEDENFILK 0.50 0.93 2.23 4.33 22.23 18.92 18.67 22.08 12.87 15.91 1.06 2.38 4.47 22.30 41.29 31.94 19.19 15.42 15.93 PPIA_VSFELFADK 0.72 1.02 2.59 4.48 22.27 48.45 37.92 28.81 14.34 14.65 CATA_GAGAFGYFEVTHDITK 1.12 0.47 1.24 2.25 8.04 80.74 46.49 79.39 46.25 25.30 CATA_GAGAFGYFEVTHDITK 1.15 0.53 1.47 2.56 8.29 75.82 48.13 67.03 45.56 24.25 CATA_FSTVAGESGSADTVR 0.62 0.90 2.27 4.79 22.02 113.39 18.95 23.42 17.60 12.70 CATA_FSTVAGESGSADTVR 0.55 0.92 2.49 4.69 22.13 113.34 41.87 33.56 18.53 11.48 CATA_NLSVEDAAR 0.30 0.69 2.24 4.95 27.12 76.51 59.91 44.70 33.85 8.64 CATA_NLSVEDAAR 0.27 0.63 2.33 4.96 27.22 79.90 61.85 41.77 30.83 12.96 B ORBITRAP C 1000 1001.52 B 10.81 C 4.09 D Sample E (1000 fmol) 11.41 9.08 11.49 10.75 11.83 65.94 29.58 17.19 29.67 17.50 90.46 186.49 1002.49 37.49 18.12 29.96 24.88 27.64 10.04 21.75 53.26 4.00 8.00 20.00 40.00 200.00 3.06 5.79 22.60 27.55 161.07 15.11 NADPH_ALIVLAHSER 1.33 3.29 8.59 18.44 88.68 17.52 7.65 13.20 10.16 17.06 CRP_ESDTSYVSLK 3.69 7.49 15.61 32.95 167.97 9.04 49.12 8.07 17.33 9.53 1.97 4.54 14.65 25.03 0.80 1.60 4.00 8.00 PPIA_FEDENFILK 0.41 0.96 PPIA_VSFELFADK 0.83 0.97 40.00 2.05 4.48 24.72 2.96 4.66 22.04 6.74 18.29 10.26 5.88 24.20 70.22 57.51 41.07 14.31 14.98 56.87 14.18 73.57 29.09 21.71 9.71 41.03 25.04 8.84 71.70 32.27 10.70 21.96 CATA_GAGAFGYFEVTHDITK 0.79 0.45 1.56 2.40 9.19 CATA_FSTVAGESGSADTVR 0.37 0.80 2.98 3.94 22.38 12.72 CATA_NLSVEDAAR 0.14 0.40 2.91 5.30 24.86 109.32 B Q-TOF C Sample A Peptide theor. fmol 140 900 120 TQ OT QTOF 21 OT TQ QTOF 6 OT QTOF 60 Inter-Lab %CV 40 Sample A (0.8 fmol) Intra-Lab %CV CATA_FSTVAGESGSADTVR NADPH_EGHLSPDIVAEQK 7 80 19 Sample A (4 fmol) CAH1_GGPFSDSYR 22 Sample A 100 22 20 TQ Sample A (20 fmol) 23 120 23 0.7 20 0.6 D E A B C Average fmol measured CAH1_GGPFSDSYR 160 D 0.5 E CV(%) 20 40 100 200 20.44 40.20 102.97 200.38 1000 3.47 2.71 3.65 1.80 14.84 4.31 19.57 139.73 792.83 7.23 30.28 32.62 27.68 4.19 90.52 212.91 1182.04 21.38 9.12 31.40 0.71 11.31 8.00 20.00 40.00 200.00 NQO1_EGHLSPDIVAEQK 3.63 7.54 18.53 35.77 179.64 58.66 23.84 16.67 13.41 NQO1_ALIVLAHSER 1.68 3.56 8.28 18.56 981.40 89.12 3.27 3.00 3.97 CRP_ESDTSYVSLK 4.86 11.05 19.15 37.98 188.45 48.40 37.45 CRP_GYSIFSYATK 2.13 4.96 14.12 24.56 148.97 16.25 18.92 0.80 1.60 4.00 8.00 40.00 21 20 fmol measured theor. fmol 25 24 180 950 fmol measured CRP_GYSIFSYATK 220 200 1000 CATA_FSTVAGESGSADTVR NADPH_EGHLSPDIVAEQK 1150 1100 1050 Sample E (40 fmol) Sample E (200 fmol) CAH1_GGPFSDSYR 1.37 986.23 934.06 29.72 theor. fmol 0 E 3.48 181.99 124.25 15.13 NADPH_EGHLSPDIVAEQK Intra-Lab %CV 5 %CV A 1.84 CV(%) 200 198.71 86.40 73.73 CAH2_SADFTNFDPR fmol measured E 100 101.79 39.73 29.31 fmol measured D Average fmol measured 40 39.82 18.22 19.66 fmol measured A 20 20.01 CAH1_VLDALQAIK CAH2_AVQQPDGLAVLGIFLK fmol measured Sample Peptide Inter-Lab %CV 9.00 0.51 0.72 theor. fmol 15 10 7.42 PPIA_FEDENFILK PPIA_VSFELFADK CAH1_GGPFSDSYR 20 %CV Sample Peptide theor. fmol CAH1_GGPFSDSYR 5 4 0 0.4 0.3 0.2 CAH1_VLDALQAIK 21.19 41.07 95.07 204.44 CAH2_AVQQPDGLAVLGIFLK 19.40 43.28 107.15 CAH2_SADFTNFDPR 20.71 43.44 4.00 theor. fmol theor. fmol 977.43 PPIA_FEDENFILK 0.51 1.14 3.16 4.65 24.84 PPIA_VSFELFADK 0.42 0.94 3.32 4.26 22.47 CATA_GAGAFGYFEVTHDITK 2.70 1.10 2.25 5.01 CATA_FSTVAGESGSADTVR 0.28 1.16 3.54 5.02 22.84 CATA_NLSVEDAAR 0.28 0.75 1.81 4.39 31.48 19 0.1 15.39 24.77 39.59 93.92 49.89 5.60 7.70 21.92 7.51 0.20 20.08 y = 0.9987x R² = 1 600 400 OT QTOF TQ OT QTOF TQ B QTOF B OT B 0.96 0.94 QTOF E OT E 1 0.9 0.8 0.4 0.2 TQ E 0.2 CAH2_AVQQPDGLAVLGIFLK 1 0.8 0.6 0.4 10 Solid bars represent the measured % coefficients of variation (%CV) between the three replicate analysis performed, averaged for all datasets. Each bar is the %CV value for the measurement of the indicated peptide. Error bars indicate the standard deviation of the values for all labs. Bars are colored according to the concentration tiers in the samples. Empty bars represent the value for Inter-Laboratory %CV for the corresponding peptide measurement. The graphs for the most concentrated sample (E) and the most diluted (A) are shown. 1.2 CAH1_VLDALQAI[K] 0.98 0.6 100 1.02 1 0.8 200 0 CAH2_AVQQPDGLAVLGIFL[K] 0.7 TQ E 0.6 QTOF E 0.5 OT E 0.4 0.3 0.2 0.92 0 0 1 0 200 400 600 theoretical fmol 800 1000 1 10 100 1000 0.9 10000 log theoretical fmol NADPH 1.2 NADPH 1000 200 NADPH_EGHLSPDIVAEQK NADPH_ALIVLAHSER 160 y = 0.8673x R² = 0.9998 120 80 y = 0.4689x R² = 0.9999 40 0.8 80 120 theoretical fmol 160 0.95 100 OT B 0.9 0.4 10 PPIA 100 log theoretical fmol TQ E QTOF E 0.85 0 1 200 TQ B QTOF B 0.6 10 1 40 CATA_FSTVAGESGSADTVR 1 0.2 0 0 CATA_FSTVAGESGSADTVR 1 NADPH_EGHLSPDIVAEQK NADPH_ALIVLAHSER log measured fmol measured fmol CAH1_VLDALQAIK 1 1000 log measured fmol measured fmol 1.2 CAH1_GGPFSDSYR CAH1_VLDALQAIK y = 1.0153x R² = 1 TQ REPRODUCIBILITY OF TRANSITION PATTERN CAH1 10000 800 QTOF Representative boxplots of the distribution of absolute concentration measurements for a peptide of each of the concentration tiers. The graphs for the most concentrated sample (E) and the most diluted (A) are shown. Measurements are grouped by type of measurement/instrument: TQ: SRM on triple quadrupole; OT, QTOF: pseudoSRM in orbitrap and Q-TOF instruments, respectively. In general, no major differences in terms of median value or dispersion were observed between the three groups. 25.76 14.23 LINEARITY OF THE RESPONSE CAH1_GGPFSDSYR CAH1_VLDALQAIK 1000 OT 3.18 7.97 97.18 0 2 TQ CAH1 1200 3 18 6.86 3.04 Tables summarizing the measurements of absolute concentration for each peptide and sample, averaged for each group of instruments, as indicated, and across all 30 datasets. The %CV of variation of the measurements are shown on the right. Red scale highlights %CV values above 20%. 0.8 1000 OT E Bar plots (left panels) represent the contribution of each of the three monitored transitions to the total MS signal, for three example peptides, for each instrument/laboratory. To compare the patterns, the dotproducts of the normalized vectors defined by the three contributions and the one corresponding to the average of all laboratories have been calculated. The plots on the right panels show the dotproduct values for each lab, grouped by type of instrument. The two peptide graphs shown on the left side are representative of the behaviour of most of the signals, showing no major differences between instruments, irrespective of the signal level (dotproducts for samples B and E shown for comparison). The plots shown on top right are an example of a peptide for which different labs selected different patterns, probably due to the presence of interfering signals. Two different patterns are clearly seen both in he bar or dotproduct plots, not depending on the instrument type. Quantification for this peptide shows accordingly a higher variability between laboratories. PPIA 30 100 PPIA_FEDENFILK PPIA_VSFELFADK PPIA_FEDENFILK PPIA_VSFELFADK 25 y = 0.5552x R² = 0.9999 20 15 log measured fmol measured fmol EUPA 2013 In order to evaluate the robustness and reproducibility, within and across laboratories, of the SRM and pseudo-SRM quantification methodology, we set up a multi-centric study (PME8) carried out at 27 laboratories, including ProteoRed-ISCIII network of proteomics facilities in Spain, several EuPA members, and other laboratories worldwide. Each participant laboratory received a set of 5 different samples A-E, prepared by spiking different amounts of the Sigma-Aldrich MSQC1 standard into a yeast lysate digest. The five samples were analyzed in triplicate by SRM or pseudo-SRM using similar chromatographic and spectrometric conditions at the different laboratories and with different instruments. Each laboratory reported results on relative quantification (fold changes between A-E samples) and absolute quantification based on the labeled peptide standards y = 0.5579x R² = 0.9993 10 10 CONCLUSIONS 1 0.1 1 10 100 The results demonstrate a good degree of reproducibility of targeted quantification measurements by SRM at different laboratories, irrespective of the method of analysis and the spectrometer used. 5 0 0 0 5 10 15 20 25 30 theoretical fmol 35 40 45 log theoretical fmol Plots of measured amounts (average of 30 datasets) in fmol versus theoretical amounts, in each of the five samples, for two different peptides of a protein of each concentration tier. Error bars represent the standard deviation of the measurements across labs. Graphs on the right show the linearity in log-log scale. A good linearity of the response was observed for all peptides in the measured ranges. A deviation from the expected theoretical amounts was observed in some cases. In the case of NADPH peptides (middle graph), both peptides give a different quantification, being one of them around half of the theoretical amount. This could be the result of an uncomplete digestion of the protein during sample preparation. In the case of PPIA (bottom graph), both peptides give the same quantification, but the value is about half of the theoretical amount. This could potentially be due to unaccuracy in the quantification of the protein standard. The average Inter-Laboratory %CV of the measured absolute protein amounts ranges from less than 10%CV for Tier 1 proteins, to 40-60% for the proteins at the lowest concentrations. The pattern of relative intensities of the transitions monitored is fairly consistent among different instruments and fragmentation modes, underscoring the utility of spectral databases for the design of quantification methods. The results obtained at each laboratory allow the assessment of the limitations in sensitivity and limits of quantification under the diverse analytical conditions used CONCLUSIONS The results demonstrate a good degree of reproducibility of targeted quantification measurements by SRM at different laboratories, irrespective of the method of analysis and the spectrometer used. The average Inter-Laboratory %CV of the measured absolute protein amounts ranges from less than 10%CV for Tier 1 proteins, to 40-60% for the proteins at the lowest concentrations. The pattern of relative intensities of the transitions monitored is fairly consistent among different instruments and fragmentation modes, underscoring the utility of spectral databases for the design of quantification methods. The results obtained at each laboratory allow the assessment of the limitations in sensitivity and limits of quantification under the diverse analytical conditions used