Chapter 4B
advertisement

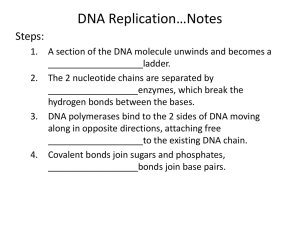
Chap. 4. Basic Molecular Genetic Mechanisms (Part B) Topics • Structure of Nucleic Acids • Transcription of Protein-coding Genes and Formation of Functional mRNA • Decoding of mRNA by tRNAs • Stepwise Synthesis of Proteins on Ribosomes • DNA Replication Goals To learn the basic mechanisms of transcription, RNA processing, translation, and replication Fig. 4.1. The Three Roles of RNA in Translation Protein translation by ribosomes requires three types of RNA (Fig. 4.17). Messenger RNA (mRNA) specifies the amino acid sequence of the protein. Each amino acid is selected based on the order of triplet codons in mRNA. Transfer RNA (tRNA) converts the information in mRNA codons into the amino acid sequence of the protein. tRNAs carry amino acids specified by the codons and base pair with the codons via their anticodons. Ribosomal RNA (rRNA) makes up the bulk of the mass of the ribosome. One rRNA species (28S rRNA) is a ribozyme that catalyzes the reaction in which the peptide bond is formed. The Genetic Code The codons for the 20 standard amino acids are specified by triplets of bases known as the genetic code (Table 4.1). Because there are 43=64 possible combinations of triplet codons, most amino acids are specified by more than one codon (degeneracy). 61 codons specify amino acids. Three do not (stop or termination codons). Termination codons tell ribosomes where to end translation of the mRNA. Most commonly, the AUG codon (specifying methionine) serves as the start codon, and tells the ribosome where to begin translation. Few deviations from the standard genetic code have been found, providing strong evidence that life on earth evolved only once. Reading of the Triplet Code There are three potential reading frames in all mRNAs. However, only one reading frame is used for translation, and is selected based on the frame in which the AUG start codon appears. Triplet codons are read in a non-overlapping, comma-less manner (Fig. 4.18). Rarely are mRNAs read in more than one frame. Likewise, frame-shifting is very uncommon. Two-step Process for mRNA Decoding Amino acids are attached in ester linkage to the 3'-terminus of tRNA, forming aminoacyl-tRNAs (Fig. 4.19, step 1). The enzymes that carry out this ATP-driven reaction are known as aminoacyltRNA synthetases. Aminoacyl-tRNA synthetases are highly accurate (high fidelity) and this helps minimize translation errors. In step 2, the amino acid is added to the growing protein chain based on codon:anticodon interactions between mRNA and tRNA. Bacteria synthesize 30-40 tRNAs, whereas eukaryotes may synthesize 50-100. Thus, a given amino acid often can be carried by more than one species of tRNA. Each aminoacyl-tRNA synthetase recognizes 1 amino acid and all of its cognate tRNAs. Structure of tRNAs tRNAs typically are 70-80 nucleotides in length. They all have a cloverleaf secondary structure and fold into an L-shaped tertiary structure (Fig. 4.20). Four double-helical stems occur, and three of these have loops of 7-8 residues at their ends. One loop (the anticodon loop) contains the anticodon. The upper stem is known as the acceptor stem and ends with a CCA sequence in all tRNAs. The amino acid is attached in ester linkage to the 2' or 3' hydroxyl group of the A residue. Many residues are modified in tRNA, and some modifications are shown in the figure. aa Codon-anticodon Base Pairing H-bonding between the 1st and 2nd positions of the codon and the 3rd and 2nd positions of the anticodon nearly always occurs via Watson-Crick base pairing. However, base pairing between the 3rd position of the codon and 1st position of the anticodon (termed the "wobble position" in both sequences) is less constrained (Fig. 4.21). For example, G, U, and I (inosine) in the wobble position of the anticodon can base pair with C/U, A/G, and C/A/U in the codon, respectively. Wobble base pairing reduces the number of tRNA genes that an organism must make to carry out translation. It also helps protect against mutations that might inactivate tRNA genes. Wobble is allowed at the codon:anticodon interaction site due to stabilization of tRNAmRNA binding by ribosomes. Ribosome Composition Ribosomes are RNA-protein supramolecular complexes. They are the most abundant type of RNA-protein complex in cells. The compositions of prokaryotic and eukaryotic ribosomes are summarized in Fig. 4.22. Although proteins outnumber rRNAs, rRNAs comprise 60% of the ribosomal mass (see Fig. 4.23). Overview of Eukaryotic Translation Initiation Like transcription, translation is mechanistically divided into initiation, elongation, and termination stages. All stages require translation factors in addition to ribosomes, mRNA, and aatRNAs. Prior to initiation of translation, the 60S and 40S subunits of the 80S eukaryotic ribosome occur in their dissociated states. As described next, the assembly of the 80S ribosome initiation complex at the start codon of the mRNA proceeds via binding of the mRNA and a charged Met-tRNAiMet initiator tRNA to the 40S subunit, with subsequent addition of the 60S subunit. Translation Initiation in Eukaryotes I Translation initiation in eukaryotes begins with three components/complexes that are shown near the top of Fig. 4.24. These are 1) the 40S ribosomal subunit, to which the eIF1, eIF1A, and eIF3 initiation factors are bound; 2) the eIF2.GTP + Met-tRNAiMet ternary complex; and 3) a circular mRNA formed by the binding of the eIF4 cap-binding complex at the 5’ end of the mRNA to poly(A) binding protein (PABP) associated with the 3’ end of the mRNA. These components associate in Steps 2 and 4 of the diagram, placing Met-tRNAiMet in the P site of the 40S subunit. Translation Initiation in Eukaryotes II In the next stage of initiation, the mRNA is scanned in the 5’ to 3’ direction until the first AUG start codon is brought into the P site (Steps 5 & 6). Then the hydrolysis of GTP by eIF2 generates a stable 48S initiation complex in which the initiator tRNA (Met-tRNAiMet) is H-bonded to the AUG codon. Translation Initiation in Eukaryotes III In the final stages of initiation, all initiation factors except eIF1A dissociate from the 48S initiation complex and the 80S subunit and eIF5B.GTP complex add on (Step 7). After eIF5B hydrolyzes GTP, the last initiation factors depart, and the stable 80S initiation complex is created (Step 8). This complex contains the complete E (exit), P (peptidyl-tRNA), and A (aminoacyl-tRNA) binding sites, with Met-tRNAiMet bound to the P site. Translation Elongation in Eukaryotes Translation elongation requires the assistance of elongation factors (Fig. 4.25). In Step 1 of elongation, the second amino acid of the polypeptide is carried to the A site of the ribosome by an EF1a.GTP complex. It binds to the mRNA via the anticodon located in the A site. In Step 2, GTP is hydrolyzed and EF1a departs. In Step 3, the 28S rRNA of the 60S subunit catalyzes peptide bond formation (see Fig. 4.17), resulting in a dipeptidyltRNA residing in the A site. In Step 4, the factor EF2.GTP binds, the ribosome translocates one codon along the mRNA, and GTP is hydrolyzed. As a result, the dipeptidyl-tRNA is placed in the P site, and the uncharged tRNAiMet enters the E site. The uncharged tRNA is ejected from the ribosome in the next cycle of elongation. Translation Termination in Eukaryotes When a stop codon (UAA, UAG, UGA) enters the A site, it is recognized and bound by the eRF1 release factor (Fig. 4.27). eRF1 forms a complex with eRF3.GTP. Hydrolysis of GTP by eRF3 results in cleavage of the linkage between the polypeptide and peptidyltRNA and release of the protein from the ribosomal post-termination complex. A protein called ABCE1 then binds to the complex, and via ABCE1 hydrolysis of ATP, the 40S and 60S subunits are separated. The 40S subunit recombines with the eIF1, eIF1A, and eIF3 factors making it ready for another round of initiation. Folding of the released polypeptide chain is aided by chaperones (not shown). Polysomes & Ribosome Recycling Polypeptide chain elongation proceeds at a rate of 3-5 amino acids per second. The efficiency of translation is increased via the binding of multiple ribosomes (polysomes) to the mRNA at a given time (Fig. 4.28b). Translation efficiency is further increased due to the complex between poly(A)-binding protein (PABP) and the eIF4-mRNA 5'-cap that occurs in mRNA (Fig. 4.28b). This circular complex positions ribosomes that have just terminated translation of the message near its 5' end. These ribosomes are recycled and rapidly reinitiate another round of translation. Mechanism of DNA Replication DNA is replicated via a semiconservative mechanism (Fig. 4.29a). In this method the parental DNA duplex separates, and each strand serves as a template for synthesis of a complementary strand. Thus the daughter DNA molecules consist of one old & one new DNA strand. The alternative conservative model for replication was ruled out based on a classic experiment conducted by Meselson & Stahl (Fig. 4.29b). DNA Synthesis at the Replication Fork An overview of semiconservative replication is presented in Fig. 4.30. The event depicted is occurring at a replication fork formed after replication has initiated at a replication origin. One strand of the lower daughter molecule (the leading strand) is being synthesized continuously in the same direction as fork movement. One strand of the upper daughter molecule (the lagging strand) is being synthesized in the opposite direction in a discontinuous manner in relatively short segments called Okazaki fragments. DNA polymerases require primers for DNA synthesis. Only one primer is needed for synthesis of the leading strand. However, each Okazaki fragment on the lagging strand is made from a primer. Primers used in DNA synthesis are composed of RNA & DNA. Eventually, RNA primers are replaced with DNA and Okazaki fragments joined together by DNA ligase. Replication of SV40 Viral DNA (Part A) The mechanism of eukaryotic replication is known mostly from the study of the replication of the SV40 virus, which infects monkeys (Fig. 4.31). SV40 is a good model system because all but one of the proteins required for its replication (viral large T-antigen) are synthesized by host cells. At SV40 replication forks, large Tantigen uses its helicase activity to unwind DNA. Both strands of single-stranded DNA are bound and coated by replication protein A (RPA) which keeps the DNA in a ideal template conformation (Fig. 4.31c). Replication of SV40 Viral DNA (Part B) The leading strand is synthesized continuously by DNA polymerase d (Pol d) (Fig. 4.31). Pol d forms a complex with replication factor C (Rfc) and proliferating cell nuclear antigen (PCNA) which keep the enzyme bound to DNA (Fig. 4.31b). RPA is displaced as the polymerase moves forward synthesizing the chain in a 5' to 3' direction. The lagging strand is synthesized discontinuously in a 5' to 3' direction from RNA/DNA primers made by a complex containing primase and Pol a. The 3' ends of primers are elongated by a second Pol d/Rfc/PCNA complex. RNase H degrades the RNA primers, and the gaps are filled in by Pol d. Nicks in the lagging strand are sealed by DNA ligase. Topoisomerase I reduces positive supercoiling ahead of large T-antigen. Bidirectional Replication of SV40 DNA Replication of SV40, and most likely all other prokaryotic and eukaryotic DNAs, occurs bidirectionally starting from a replication origin. Bidirectional replication increases the rate at which DNA molecules are copied. The bidirectionality of replication has been demonstrated in experiments such as shown in Fig. 4.32. When a mixture of replicating SV40 DNA molecules are linearized by cutting with a restriction enzyme, the replication bubbles observed all are centered at the same position on the DNA. This indicates replication has proceeded in both directions from the origin. Model for Bidirectional DNA Replication A conceptual model for initiation of bidirectional replication and fork movement away from a replication origin is shown in Fig. 4.33. For SV40, large T-antigen unwinds the parental strands. In eukaryotic chromosomal replication, cellular helicases known as MCM proteins perform unwinding. Each eukaryotic chromosome contains multiple replication origins separated by tens to hundreds of kilobases. The activation of MCM helicases (and thereby, DNA replication) is controlled by S-phase cyclindependent kinases (Chap. 19).