Document
advertisement

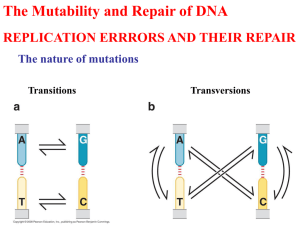
(Genotype) Replication (Chemistry, Phenotype) Replication (Chemistry, Phenotype) DNA Replication Action of DNA polymerase. DNA polymerases assemble incoming deoxynucleoside triphosphates on single-stranded DNA templates such that the growing strand is elongated in its 5’ to 3’ direction. Priming of DNA synthesis by short RNA segments. DNA Polymerase DNA polymerase activity 3’-5’ exonuclease 5’-3’ exonuclease 3’-5’ exonuclease cleaves misincorporated nucleotides 5’-3’ exonuclease Cleaves RNA primers from growing strands 1) Helicase: separate DNA strands 2) DNA primase: RNA polymerase that lays down the primer 3) DNA polymerase replicates DNA 4) b clamp increases processivity 5) DNA gyrase/topoisomerase: prevents supercoiling Helicase and gyrase/topoisomerase E. coli DNA replicated at 1000 nucleotides/s 1300 µM DNA would flail around at 100 revolutions/s Since it is circular it instead supercoils Accumulates +100 supercoils/s these need to be alleviated DNA is naturally negatively supercoiled and this promotes unwinding However, GyrA, type II topoisomerase further helps by introducing negative supercoiling Proofreading E. coli polymerase makes mistakes every 109 -1010 nucleotides added Given that the genome is 4.6x106 bp this means 1 error per 1000 to 10000 replications Proofreading requires Proper base pairing Even a properly inserted nucleotide is removed a significant amount of the time just to be sure…. Many types of DNA polymerase in E. coli Pol I: DNA repair, excision of RNA primers Pol II: DNA repair Pol III: main DNA replicase Pol IV and V? Eukaryotic DNA polymerases Pol : a primase Pol : main polymerase Pol : mitochondrion Pols b: DNA repair The reactions catalyzed by reverse transcriptase. NMR structure of the telomeric oligonucleotide d(GGGGTTTTGGGG). Errors do occur Point mutations Insertions Deletions Rearrangements Spontaneous mutation rate = 10-10 to 10-12/cell division in bacteria 1 nucleotide change/per 1000 takes 200,000 yrs. in humans Types and sites of chemical damage to which DNA is normally susceptible in vivo. Red, oxidation; blue, hydrolysis; green, methylation. Mispairing of A with C or G with T: Tautomerization H O O H N N A N H O N H N N G T H N N A N N H N H2N N N H N C* N N O C N O T* N N H O H N H H N N N N O Hydrolysis Need to get rid of uracil in DNA Hydrolysis Depurination occurs 6000 times per day in a mammalian cell Methylation Oxidation O N O N ¥OH NH NH HO N H N H NH2 N H N NH2 Guanosine O O H N N HO HO H NH NH N H N NH2 N H N NH2 Hydroxoguanosine Oxidation P base O O H P ¥OH H base O O H H H H H O P P P O O O H O H H O CH3 O H H H H H H O P Strand Breaks H Deletions due to strand breaks Insertions and deletions Caused by repetitive sequences and intercalators abasic sites single strand breaks Triplet repeat expansion/contraction CAG or CGG repeats (polyGln) Triplet repeat expansion/contraction CAG or CGG repeats (polyGln) Deletions Six main types of repair mechanism Direct repair Mismatch repair Base Excision Repair Nucleotide Excision Repair SOS Repair Homologous recombination Direct repair: photolyase Direct repair: O6-methyl guanosine demethylase Mismatch repair Points out into major groove H3C N N N H H N N Base Excision Repair Nick translation by DNA pol I Nucleotide Excision Repair SOS Repair/Translesion synthesis/Error prone polymerase When you hit a lesion DNA pol III falls off It is replaced by pols without proofreading this causes the polymerase to pass over the lesion without changing it This, of course, introduces errors but it is better than no replication at all So stress induces deliberate mutation! Why? To enhance evolvability, perhaps? One of the best ways to repair DNA is to use homologous recombination: sharing information between homologous chromosomes Homologous end-joining Gross Rearrangements Transposon and transposable elements DNA only transposons Retroviral like retrotransposons Nonretroviral retrotransposons Transposons are ‘jumping genes’ that make use of recombination to jump around Chromosomal rearrangement via recombination. (a) The inversion of a DNA segment between two identical transposons with inverted orientations. Chromosomal rearrangement via recombination. (b) The deletion of a DNA segment between two identical transposons with the same orientation.