Data Mining and Marker Validation - University of California, San
advertisement

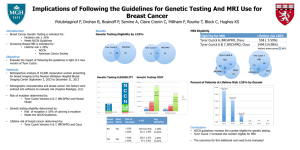
A systems approach to marker guided therapy in breast cancer Joe W. Gray, Ph.D. Lawrence Berkeley National Laboratory University of California, San Francisco A systems approach to marker guided therapy in breast cancer Breast cancer overview and statement of the problem An in vitro systems approach to match treatment to “ome” Improving and testing the model A systems approach to marker guided therapy in breast cancer Breast cancer overview and statement of the problem An in vitro systems approach to match treatment to “ome” Improving and testing the model Stage Distribution and 5-year Relative Survival by Stage at Diagnosis for 1999-2006, All Races, Females Stage at Diagnosis Localized (confined to primary site) Regional (spread to regional lymphnodes) Distant (cancer has metastasized) Unknown (unstaged) Stage Distribution (%) 5-year Relative Survival (%) 60 98.0 33 83.6 5 23.4 2 57.9 World wide incidence - 1,150,000/yr Worldwide mortality - 410,000/yr SEER Registry Overall goal Improve treatment by identifying molecular subtype markers that • predict resistance to existing therapies • predict response to experimental therapies Hundreds of compounds are approved or well along in the developmental pipeline How do we find the most effective for breast cancer? International cancer genomics efforts are substantially increasing the number of recognizable cancer subtypes that may respond differentially to specific therapies State of the breast cancer genome • Remarkable genomic and epigenomic heterogeneity between and within tumors • Hundreds of genes and gene networks are deregulated in ways that contribute to cancer pathophysiology • Subtypes are defined by aberrations at multiple levels: mutation, structure, copy number, chromatin modification, ncRNA, … • Subtypes defined by recurrent aberrations are associated with outcome • Response varies with subtype State of the breast cancer genome • Remarkable genomic and epigenomic heterogeneity between and within tumors • Hundreds of genes and gene networks are deregulated in ways that contribute to cancer pathophysiology • Subtypes are defined by aberrations at multiple levels: mutation, structure, copy number, chromatin modification, ncRNA, … • Subtypes defined by recurrent aberrations are associated with outcome • Response varies with subtype State of the breast cancer genome • Remarkable genomic and epigenomic heterogeneity between and within tumors • Hundreds of genes and gene networks are deregulated in ways that contribute to cancer pathophysiology • Subtypes are defined by aberrations at multiple levels: mutation, structure, copy number, chromatin modification, ncRNA, … • Subtypes defined by recurrent aberrations are associated with outcome • Response varies with subtype How do we make the optimal match between drug and subtype? A systems approach to marker guided therapy in breast cancer Breast cancer overview and statement of the problem An in vitro systems approach to match treatment to “ome” Improving and testing the model We use a collection of 50+ breast cancer cell lines to model the molecular diversity of primary tumors • Therapeutic approaches can be tested quickly to identify subtype specific responses • Model can be characterized at great molecular depth to identify predictive markers • Model can be manipulated to test predictions To what extent do the cell lines represent what we know about breast cancer? Cell lines model gene expression subtypes, recurrent copy number chances and mutations We have assessed ~100 therapeutic strategies in 50 cell lines Emphasis on signaling pathways Establishing associations between response and molecular subtypes UCSC Cancer Genome Browser Molecular features Biological features Approximately half of compounds tested show significant molecular subtype specificity A Luminal Bas al Claudin-low Normal Ambiguous GI50 GI50Relative cell 1e-06 AKTi We are especially interesting in identifying 1e-07 genomic drivers for molecular response Log drug concentration CI1040 1e-05 GI50 1e-04 Associations number at 3 days 1e-05 Lapatinib 1e-06 1e-05 1e-07 Associations 1e-06 1e-08 Cell line Cell line Kuo, Guan, Hu, Bayani 2007 Most effective targeted agents are linked to genomic markers that predict response Imatinib mesylate CML Imatinib mesylate Sunitinib Nilotinib Dasatinib GIST Dermatofibrosarcoma protuberans Hypereosinophylic syndrome Melanoma Trastuzumab Pertuzumab Lapatinib Breast Gefitinib, Erlotanib Cetuxumab Lung cancer Bowel PKC412, SU11248, CMT53518 AML, ALL PARP inhibitors Breast Ovarian PLX4032 Melanoma Crizotinib Lung Tamoxifen, AIs Breast cancer BCR-ABL translocation Oncogene addiction (1982) c-KIT mutation PDFGR mutation Oncogene addiction (1999) HER2 amplification Oncogene addiction (1985) EGFR mutation Oncogene addiction (2004) FLT-3 mutation, tandem duplication Oncogene addiction (1996) BRCA1/2 mutation Synthetic lethality (2005) BRAF mutation Oncogene addiction (2002) EML-4 ALK translocation Oncogene addiction (2007) ER expression Lineage (1800s) *Except VEGFR and proteosome inhibitors ~25% of compounds are significantly associated with genome copy number abnormalities Spellman, Sadanandam, Kuo Kuo, Spellman, Sadanandam Platinum, anti-metabolites and antimitotic apparatus protein inhibitors effective in basal subtype cells Luminal Basal Claudin-low Sensitive Resistant PI3K inhibitor PI3K inhibitor PI3K inhibitor AURK inhibitor PLK1 inhibitor Response to mitotic apparatus inhibitors is associated with transcriptional upregulation of a network of mitotic apparatus genes Mao, Hu et al Why does this network exist? Expression of mitotic apparatus genes is associated with amplification of transcription factors that target mitotic apparatus genes Christina Clark, Carlos Caldas All genes in the mitotic apparatus signature are targeted by these transcription factors Mao, Curtis, 2010 Kuo, Spellman, Sadanandam EGFR, ERBB2, PI3K inhibitors, HDACs effective in luminal subtype cells Luminal Basal Claudin-low PI3K inhibitor PI3K inhibitor PI3K inhibitor AURK inhibitor PLK1 inhibitor Hierarchical clustering of 31 significant subtype specific drugs and BrCa cell lines. Luminal subtype preference for ERBB2 and AKT pathway inhibitors “explained” by the subtype specificity of activating genomic aberrations X GI50 Lapatinib GI50 AKTi PIK3CA mutation PTEN mutation PIK3CA PTEN Aberrations interact - AKT inhibitors synergize with lapatinib in ERBB2+, PIK3CAmt cells Synergistic Antagonistic? PIK3CA mutation (blue is mutant) Drug combination dose H1047R PIK3CA, E307K PTEN K267fs*9 PTEN H1047R PIK3CA E545K PIK3CA H1047R PIK3CA E545K PIK3CA K111N PIK3CA Korkola, Cooper, et al 2010 A systems approach to marker guided therapy in breast cancer Breast cancer overview and statement of the problem An in vitro systems approach to match treatment to “ome” Improving and testing the model Complicating factors • Microenvironment • Response not durable • Response heterogeneity The microenvironment modulates response to ERBB2 targeted drugs AU565 ERBB2 amp SKBR3 ERBB2 amp 2D monolayer 3D matrigel HCC1569 ERBB2 amp BT549 ERBB2 norm Wiegelt, et al., Breast Cancer Res Treat 122:35–43, 2010 The microenvironment modulates the signaling network HER3 HER2 PI3K a,b,g,d IRS1 PDK1 RAF Akt MEK TSC2 MAPK Rheb PRAS40 mTorC2 mTorC1 PKCα S6K1 S6 Inhibition of microenv. signaling also should modulate response HER3, PDK1, Akt, … COX2, CREB, cJun, NFkB, ATF2, ER, Tcf/Lef, Rb, AP1, cFos, CXCR4, ETS, HIF1a, MYC -> CBX5 Inhibition of b1-integrin signaling enhances response to ERBB2 targeted drugs in 3D but not 2D AU565 ERBB2 amp SKBR3 ERBB2 amp AIIB2 None HCC1569 ERBB2 amp BT549 ERBB2 norm Wiegelt, et al., Breast Cancer Res Treat 122:35–43, 2010 Microenvironment dependent response may explain why treatment of metastatic disease is difficult Can we identify microenvironment independent therapies? This motivates assessment of pathway function in situ Britt Marie Ljung TOF-SIMS “ome” imaging Immunohistochemistry or in Primary Ion Beam situ hybridization with mass tag labeled reagents. Each tag is a color. Tag 1 Map Total Area Spectrum Tag 2 map m/z Sample More complications ERBB2 inhibition is not durable Amin et al, Science TM 2010; 2: 16ra7. Understanding response dynamics HER3 HER2 PI3K a,b,g,d IRS1 PDK1 RAF Akt MEK TSC2 MAPK Rheb PRAS40 Mills, Moasser et al mTorC2 mTorC1 PKCα S6K1 S6 HER3, PDK1, Akt, … COX2, CREB, cJun, NFkB, ATF2, ER, Tcf/Lef, Rb, AP1, cFos, CXCR4, ETS, HIF1a, MYC -> CBX5 Statistical and dynamic modeling to understand long term behavior Center for Cancer Systems Biology – – ODE model for short term effects (Soulaiman Itani) A hybrid Boolean-ODE model Signaling occurs in 3 dimensions using to model longer term effects[Chen 2009] (YoungNetwork behavior is context dependent Hwan Chang) Need to understand the emergent properties of complex, cross coupled systems Tomlin lab Molecular responses are heterogeneous – a partial explanation for lack of durability? Digital v. analogue drug responses Sorger et al A systems approach to marker guided therapy in breast cancer TCGA/ICGC projects are defining a growing number of distinct subtypes In vitro systems suggest at least half of all therapeutic compounds show subtype specificity Improving the model - Modeling the microenvironment, heterogeneity and long term durability Collaborators Clinical science (I SPY etc) Laura Esserman (UCSF) Laura Van’t Veer (UCSF) Rick Baehner (UCSF) Nola Hylton (UCSF) John Park (UCSF) Hope Rugo (UCSF) Britt Marie Ljung (UCSF) Hubert Stoppler (UCSF) Fred Waldman (UCSF) Mitotic apparatus networks Zhi Hu Jian Hua Mao Shenda Gu Barbara Weber (GSK – then) Richard Wooster (GSK) Christina Clark (Cambridge) Carlos Caldas (Cambridge) Cell line system biology Wen-Lin Kuo Jim Korkola Nick Wang Nora Bayani Brian Cooper Mara Jeffress Anna Lapuk Demetris Iacovides Mina Bissell Martha Stampfer Terry Speed (UCB) Claire Tomlin (UCB) Michael Korn (UCSF) Frank McCormick (UCSF) Gordon Mills (MDACC) Yiling Lu (MDACC) Peter Sorger (Harvard) Genome biology Paul Spellman Anguraj Sadanandam Laura Heiser Shannon Dorton Jing Huang Steffen Durinck Obi Griffith Lakshmi Jakkula Francois Pepin Andy Wyrobek David Haussler (UCSC) Josh Stuart (UCSC) Project management Heidi Feiler Shradda Ravani NCI Center for Cancer Systems Biology, The Cancer Genome Atlas, CPTAC, Bay Area Breast Cancer SPORE, Atwater foundation, GSK, Roche, Millenium, Pfizer, Progen, Cytokinetics, Cell Biosciences, DOD Innovator, SU2C