HardsisonDynamEpiGen_BCN
advertisement
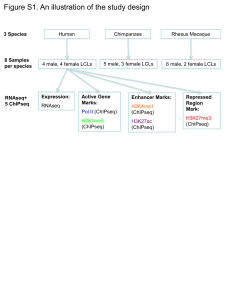
Dynamics of epigenetic states during erythroid differentiation Ross Hardison July 20, 2010 Barcelona Investigators on one mouse ENCODE project • • Cal Tech - Barbara Wold Penn State – – – – – – – – • Hardison Stephan Schuster Frank Pugh Robert Paulson Francesca Chiaromonte Yu Zhang Webb Miller Anton Nekrutenko Childrens’ Hospital of Philadelphia – Mitch Weiss – Gerd Blobel • Emory Univ. – James Taylor • Univ. Massachusetts – Job Dekker • Duke Univ. – Greg Crawford, consultant – Terry Furey, consultant Hematopoiesis Erythroid differentiation GATA1, partners, teammates Somatic cell model to study GATA1 function in vitro hematopoietic differentiation Gata1– ES cells erythropoietin stem cell factor immortalize G1E Erythroid progenitors BFU-e, CFU-e add back GATA-1, hybrid protein with ER G1E-ER4 estradiol G1E-ER4+estradiol Differentiated erythroblasts Changes in transcription factors and cell morphology during eythroid differentiation Changes in gene expression during erythroid differentiation Features interrogated by ChIP-seq and RNAseq assays DNase hypersensitive sites CTCF ChIP-seq finds previously known distal CRMs Known CRMs DHSs GATA1 TAL1 CTCF H3K4me1 H3K4me3 PcG: H3K27me3 + + + + + + + - = no GATA1 + = GATA1-ER activated GATA1 activates Zfpm1 by displacing GATA2 and retaining TAL1 Known CRMs DHSs + GATA1 + TAL1 + - GATA2 H3K4me1 + + - H3K4me3 + - PcG: H3K27me3 + - GATA1 represses Kit by displacing GATA2 and TAL1 Known CRMs DHSs GATA1 TAL1 GATA2 H3K4me1 H3K4me3 PcG: H3K27me3 + + + + + + + - GATA1 action via TAL1 is common but not universal Chromatin states from integrative analysis: limited change during differentiation chromHMM segmentations: Jason Ernst and Manolis Kellis, MIT Broad GATA1-occupied segments of DNA: Vast majority are in active chromatin, few change states Dominant mark H3K4me3 H3K4me3 H3K4me1 H3K27me3 None H3K4me1 H3K27me3 None Changes in chromatin state at Btg2 Quantum change in chromatin state distinguishes expressed from non-expressed loci Chromatin state precedes GATA1-induced TF changes Chromatin state established (mostly): Active Repressed Dead zones Induction and repression: Dynamics of transcription factor binding within the alreadyestablished chromatin context. Chromatin condenses Nucleus removed Thanks Weisheng Wu, Yong Cheng, Demesew Abebe, Cheryl Keller Capone,Ying Zhang, Ross, Swathi Ashok Kumar, Christine Dorman, David King ….Tejaswini Mishra, Nergiz Dogan Collaborating labs: Mitch Weiss and Gerd Blobel (Childrens’ Hospital of Philadelphia), James Taylor (Emory) Webb Miller, Francesca Chiaromonte, Yu Zhang, Stephan Schuster, Frank Pugh, Bob Paulson (PSU), Greg Crawford (Duke), Jason Ernst, Manolis Kellis (MIT) Funding: NIH NIDDK, NHGRI (ARRA), Huck Institutes of Life Sciences and Institute for Cyberscience, PSU Big changes at Hbb-b1 Known CRMs DHSs GATA1 TAL1 Pol2 H3K4me1 H3K4me3 PcG: H3K27me3 + + + + + + + - TAL1 occupancy, ER4 GATA1 causes TAL1 to move at some CRMs, but retains it at others TAL1 occupancy, ER4 TAL1 occupancy, G1E GATA1 occupancy, ER4 All GATA1-occupied segments active as enhancers are also occupied by SCL and LDB1 Tripic et al. (2009) Blood Distinguishing genes induced vs repressed by GATA1 Feature Induction Repression GATA1 occupancy and transcription start site Close Distal Multiple GATA1 occupied segments per gene More (58% of genes) Fewer (24% of genes) GATA1 and TAL1 co-occupancy Almost always Subset has GATA1+ TAL1- Binding site (WGATAR) is under constraint More frequently Less frequently Polycomb mark around TSS Never Subset has Pc Kinetics of GATA1-regulated Gene Expression GATA1-induced (>2-fold) 1048 genes • known targets • new gene discovery GATA1-repressed (>2-fold) 1568 genes • stem cell/progenitor markers • proto-oncogenes (Kit/Myc/Myb) • function unknown Affy 430 2.0 Globin RNA dominates the transcription profile RNA-seq Change in expression Swathi A. Kumar, Tejaswini Mishra, et al. Induction and repression occurs within active chromatin Histone modification profiles across TSS+/ 4kb High Level of expression Low H3K4me3 at TSS correlates with expression level