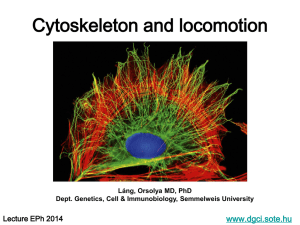
an actin cytoskeleton
advertisement

5. Cell adhesion and the actin
cytoskeleton
The cell cortex
The cytoskeleton networks
The actin cytoskeleton
Adhesion proteins
The extracellular matrix
The cell architecture is regulated by the adhesion zones
1
The cell cortex
Actin microfilaments
0.2 µm
Protein complexes
5 nm
membranes
Medalia et al. 2002
Science 298: 1209-1213
2
100 nm cryoelectron tomography
The cell cytoskeleton networks
Cells possess two or three cytoskeleton networks made of polymerized proteins :
an actin cytoskeleton (microfilaments), a tubuline one (microtubules) and
intermediate filaments
The cycle of microfilament polymerisation-depolymerisation is coupled to ATP
hydrolysis by actin
The cycle of microtubule polymerisation-depolymerisation is coupled to GTP
hydrolysis by tubulin
Intermediate filaments are controlled by protein phosphorylation and
dephosphorylation
About hundred proteins control the growth and dissociation rates or are associated
to microfilaments and microtubules : actin binding proteins, microtubule associated
Intermediate filaments
proteins, bundling proteins, molecular motors ...
3
microfilaments
microtubules
The actin cytoskeleton
Microfilaments are actin polymers, an ATP binding and ATP hydrolysing
protein. The cycle of microfilament polymerisation-depolymerisation is
coupled to ATP hydrolysis by actin
Microfilaments constitute the membrane cortex which allows the deformation
of the plasma membrane. Stress fibers are actin microfilaments that link
adhesion focal points. Actin microfilaments are reversibly linked to the
plasma membrane by specific proteins (talin, catenin, ezrin ...)
Mechanical forces are exerted by actin polymerization at the plasma
membrane and between actin microfilaments by molecular motors
reflection interference contrast microscopy
actin immunofluorescence
4
Structure of actin microfilaments
barbed
end
pointed
end
4 nm
5
Actin polymerization kinetics : 1) no hydrolysis
At steady state,
V = (1/a)dL/dt = konC – koff
Critical concentration :
Cc = koff/kon = Keq
Cc+end = Cc-end
V
+ end
(polymerization
rate)
- end
Cc
C
(monomer
concentration)
6
Actin polymerization kinetics : 2) ATP hydrolysis
V
+ end
(polymerization
rate)
- end
AATP
AADP
CcATP
+ end
- end
C
CcADP (monomer
concentration)
ATP hydrolysis
Pi
2 sec
At steady state, during treadmilling
V = VT(+) + VD(-) = konT.CT – koffT + konD.CD – koffD
CD ~ 0.1 CT , koffT < koffD and konD < konT
In steady state CTc koffD/konT
7
Experimental polymerization dynamics
Fujiwara et al. PNAS 2007 104 : 8827–8832
CTc koffD/konT 0.25/11.6 = 0.02 µM
Physiological actin concentration : about 100 µM
Most actin is complexed to thymosin [ActinT] 10 µM
The theoretical actin microfilament elongation rate is :
dL/dt = a.VT = a.konT.[ActinT] < (4 nm).(12 µM-1.s-1).(10 µM) = 0.48 µm.s-1
At the pointed end :
-dL/dt = a.VD = a.koffD = (4 nm).(0.25 s-1) = 1 nm.s-1
To be compared to the actual velocity of cell edge movements : up to 10 µm.s-1
8
In vivo evidence of microfilament
treadmilling by FRAP experiments
Injection of
rhodamineactin
Localized
photobleaching
Transport
(treadmilling )
9
How cytoskeleton molecules exert forces ?
1. Actin polymerization : the elastic brownian ratchet model
(Mogilner and Oster 1996 Biophys. J. 71 : 3030-45)
Stall force
Mechanical work = ATP hydrolysis
F.d = DG/N = DG° + RT ln{[actinATP]}
= kBT ln{kon[actinATP]/koff}
actinATP + filamentN filamentN+1 + Pi
10
How cytoskeleton molecules exert forces ?
2. Actin microfilament pressure at nucleation zones
(C Sykes & J Prost (2005)
P.N.A.S., 102, 7847-52)
Actin nucleation at the bead surface
Concentric growth of actin
microfilaments
Shear stress accumulation
Breaking the actin gel, resulting in
asymmetric growth
11
Visualization of F-actin network movement in motile keratocytes with FSM
Yam et al. JCB 178:1207-1221, 2007
30x real time
Actin filament dynamics
12
12
How cytoskeleton molecules exert forces ?
3. Molecular motors sliding on microfilaments
13
Regulation of cytoskeleton dynamics
↑ Stress fibers
↑ Lamellipodia
Ridley & Hall 1992
Allen et al. 1997
↑ Filopodia
14
Example : microvilli at the plasma membrane
Brush border cells of the intestinal epithelium
Plasma membrane
Actin cytoskeleton
15
Cell adhesion and the actin
cytoskeleton
The cell cortex
The cytoskeleton networks
The actin cytoskeleton
Adhesion proteins
The extracellular matrix
The cell architecture is regulated by the adhesion zones
16
The extracellular matrix and cell adhesion molecules
The extracellular matrix is made of
macromolecules (proteins and
polysaccharides) synthesized and
secreted by cells. It provides a
specific mechanical and chemical
environment for the cells.
Cell adhesion molecules are proteins
expressed at the surface of cells that
mediate binding to other cells or to
the extracellular matrix.
17
Extracellular matrix proteins have a complex structure
Proteins of the ECM
have a very large multidomain structure
They often contain
growth factor-like
domains or bind growth
factors
A) Fibronectin. Encoded by a single gene but alternatively spliced at three regions [blue circles and box and V (variable) segment] to generate 12 proteins in
rodents and 20 in humans. FN3 domains are widespread in ECM proteins. Binding sites for other matrix proteins are marked. The heparan sulfate–binding site can
interact with proteoglycans or with syndecan, an integral-membrane proteoglycan. Integrin-binding sites; RGD (indicated by an asterisk) and LDV (Leu-Asp-Val,
indicated by a pound sign). FN is a proangiogenic molecule, whose function depends on both the RGD site and the two alternatively spliced FN3 domains. FN also
binds the proangiogenic growth factors VEGF and HGF.
B) Fibrillin-1. Fibrillins include EGF-like domains, found in many ECM proteins, as well as TB (TGFb-binding, denoted by T) and hybrid (H) domains, specific to
fibrillins and LTBPs. Binding sites for other matrix proteins and growth factors are marked.
C) LTBP-1. Four-gene family with structures related to fibrillins. Known binding sites for TGF-b/LAP latent complex (SLC, blue), fibrillin, and FN are marked. RGD
(asterisk) sequences in fibrillins and LTBPs may bind integrins.
D) Thrombospondin-1 (TSP-1). TSPs contain TSP1 repeats (also found in other ECM proteins), EGF-like repeats, and a VWC domain, known in other proteins to
bind BMPs. TSP3 repeats (purple) and C-terminal domains are unique to TSPs and bind multiple Ca 2+ ions. The RGD (asterisk) sequence is known to bind to
integrins. TSPs 1 and 2 have the structure shown, and both have antiangiogenic activity located in the TSP1 repeats, which bind to the CD36 receptor (39)
RO Hynnes (2009) the extracellular matrix : not just pretty fibrils Science 326 : 1216-19
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
Fibrillin (2 µm)
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
Cell adhesion molecules and cell adhesion structures
20
Classes of cell adhesion molecules
Some family
members
Ca2+ or Mg2+
dependence
Type
Cytoskeleton association
Cell
structures
Cell-Cell Adhesion
Classical cadherins
E, N, P, VE
yes
homophilic
actin filaments (via catenins)
adherens
junctions
Desmosomal cadherins
desmoglein
yes
homophilic
Intermediate filaments (via
desmoplakin, plakoglobin, and
other proteins)
desomosomes
Ig family members
N-CAM
no
both
unknown
no
Selectins (blood cells
and endothelial cells
only)
L-, E-, and
P-selectins
yes
heterophilic
actin filaments
no
Integrins on blood cells
αLβ2 (LFA-1)
yes
heterophilic
actin filaments
no
many types
yes
heterophilic
actin filaments (via talin, filamin, αactinin, and vinculin)
focal adhesions
α6β4
yes
heterophilic
intermediate filaments (via plectin)
hemidesmosome
s
syndecans
no
heterophilic
actin filaments
no
Cell-Matrix Adhesion
Integrins
Transmembrane
proteoglycans
21
Madin-Darby Canine Kidney cells, an epithelium model
Polarized cell : two compartments
around the cell
22
Tight junctions separates apical from basolateral compartments
The sealing strands hold adjacent membranes together.
They are composed of transmembrane proteins
(claudins, occludins) that make contact across the
intercellular space and create a seal.
Electron micrographs of cells in an
epithelium in which a small, extracellular,
electron-dense tracer molecule has been
added to either the apical side (on the left) or
the basolateral side (on the right). In both
cases, the tracer is stopped by the tight
junction. (photo Daniel Friend)
23
The cell architecture is regulated by the adhesion zones
http://www.cytoo.com/
24
25
Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity.
Thery M, Racine V, Piel M, Pepin A, Dimitrov A, Chen Y, Sibarita JB, Bornens M.
Proc Natl Acad Sci U S A 103(52):19771-6. 2006.
Golgi apparatus (in red) in RPE1 cells in
standard culture conditions
Golgi apparatus (in red) in RPE1 cells on
26
fibronectin micropatterns (in green)