BIO315
advertisement

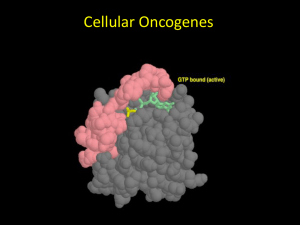
Objectives for protein section II • Explain conformational changes in proteins. • Explain how GTP binding & hydrolysis allows ras & other G-proteins to function as molecular on/off switches. • Discuss the relationship of ras normal function, ras mutants, & ras’ role in cancer. • Predict how amino acid substitutions in a protein will affect its structure and function. • Define protein families and discuss them in terms of gene duplication, evolution, and functional constraints. • Compare and contrast protein domains and protein motifs. Ras protein & the relationship of form & function • • • • • Protein = single polypeptide 189 amino acids 21 kD (kiloDalton) Relays signals in cytoplasm “Small” G-protein = Guanine nucleotide binding protein • Involved in cancer as a protooncogene Ras is a molecular on/off switch GDP GTP Exchange SOS GDP GTP Off On Hydrolysis raf on Pi mitosis Ras undergoes a conformational change when bound to GTP Conformational change = a predictable change in protein structure that is associated with biological activity. Conformational change could be due to the binding of another protein, a certain nucleotide, the addition of a phosphate, etc… Ribbon Cartoon of Ras with GTP Guanine Base Ribose sugar 3 phosphates switch II switch I g -phosphate can be hydrolyzed From C. Branden & J. Tooze. 1999. Introduction to Protein Structure, 2nd Ed. Ras conformational changes upon GTP binding (switch I, switch II) GDP GTP sII sI With GTP, more of switch I becomes b sheet. This b strand forms a b sheet with 2 strands in raf. The preceding function of ras is required for the proper function of all NORMAL mammalian cells. Ras only participates in the development of cancer when the gene for ras is mutated (an oncogene) and the resulting ras protein functions incorrectly (an oncoprotein). Mutated ras is involved in cancer • 25-30% of all human cancers contain mutated ras • mutant gene causing cancer = oncogene • resulting mutant protein causing cancer = oncoprotein • Oncogenic mutations in ras result in SINGLE AMINO ACID CHANGES • ras leads to cancer when it is overactive Mutations in GTP binding region disrupt hydrolysis and create oncoproteins Essential glycines (gly-12) required to position GTP Glutamine-61 required for GTP hydrolysis. Links to resources and tutorials on ras and protein structures • Chime tutorial on ras structure and oncogenic mutations – http://webhost.bridgew.edu/fgorga/ras/default.htm • A cartoon movie of ras conformational changes – http://bioinfo.mbb.yale.edu/MolMovDB/cgibin/morph.cgi?ID=55051-2222 • Basic tutorial on protein structure with G protein (good for alpha helix & beta sheet) – http://info.bio.cmu.edu/Courses/BiochemMols/ProtG/ ProtGMain.htm Ras is part of a protein family • Protein family = similar, but not identical proteins within an organism. • Proteins within a family have similar AA sequences, and therefore, similar (but generally not identical) structure and function. • Protein families are created by duplication of an ancestral gene. • The genes then diverge over time, potentially developing distinct, specific functions. Mammals have three different Ras genes and proteins • H-Ras – most abundant in skin. • K-Ras – most abundant in gut and thymus. • N-Ras – most abundant in testis and thymus. Ras is also part of G-protein superfamily • Other G-proteins – – – – – Rho Rac Rap1 Ran Arf Unrooted tree based on AA sequence Rap1A H-ras Rac1 K-ras RhoA N-ras, M N-ras, H Ran Arf1 In Class Problems 1. Explain why rap1A is located closer to the ras proteins than the other proteins are. Rap1A binds to raf and is involved in the same signaling pathways, so you would expect its structure to be most similar to ras. 2. Explain the grouping of rac1 and rhoA together on a branch. Rac1 and rhoA are both involved in signaling via cadherins and the actin cytoskeleton. They share the most functions, so they are the most similar to each other. 3. Explain the grouping of ran and arf1 together on a branch. Ran and arf1 share somewhat similar functions in that they are both involved in transport within the cell, so they are similar. However, their functions are very distinct, & so are their sequences. 10 30 20 40 Green = 100% identical 50 60 70 80 100 90 Yellow = many identical Blue = similar 110 120 130 140 150 #s = AA # based on human N-ras 160 170 180 In Class Problem • Refer to the sequence alignment in the preceding figure. Which nine amino acids are identical between all the proteins shown? Write their number and single letter code. Predict the area where these nine amino acids would be located in the tertiary structure of ras. gly-10, gly-15, lys-16, thr-35, asp-57, gly-60, lys-117, asp-119, ala-146 The one function that all of these proteins share is the binding of GDP/GTP; therefore, you would predict that the most commonly shared AAs would be those involved in binding the nucleotide. The 9 completely conserved AAs (green) in our G protein examples are located precisely around the nucleotide (blue) and most bond with it. C N Rotated ~180o Switch II Switch I Movie available on Blackboard site under Course Documents