2011_InstructorSlidesR
advertisement

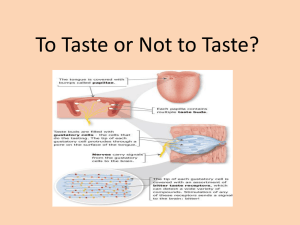
DNA Sequencing Research Projects for Summer Programs or Undergraduate Courses Julie A. Emerson Department of Biology Amherst College Amherst, MA 01002 Objectives of Group Research Projects • Enable participants to use the polymerase chain reaction (PCR) and DNA sequence analyses to discover something new about themselves or the surrounding microbial environment • Run 2-3 different projects, to keep group number to a manageable size and so different groups can present to and learn from each other • Select genes for study that have easily-identifiable differences in DNA sequence in the test population, so that comparisons can be made between test subjects • For projects using human subjects, select genes that are associated with interesting human traits • Avoid selecting genes that are associated with human diseases/disorders or that may raise questions about paternity • The time frame of the projects, from introduction to final presentations, must fit within a 10-12 day window for summer program, or in several 2-4 hour lab periods for a course Four projects have been designed and implemented since 2005: Project #1 – Molecular Identification of Human Polymorphisms in the Melanocyte-Stimulating Hormone Receptor (MC1R) Gene Project #2 – Adult Lactase Persistence and Associated Polymorphisms in the Human Lactase Phlorizin Hydrolase Gene Project #3 – Identification of Single Nucleotide Polymorphisms in a Human Bitter Taste Receptor Gene Project #4 – Molecular Identification of Bacteria in the Environment Group Research Projects #1-3 : Molecular Identification of Human Single Nucleotide Polymorphisms Day 1: Volunteers fill out Human Subjects Questionnaire ↓ DNA isolation from human cheek cells ↓ PCR using MC1R, lactase or PTC forward and reverse primers ↓ Day 2: Analytical agarose gel of PCR products ↓ Day 3: Spin purification of PCR products ↓ Analytical agarose gel of purified PCR products and quantification of DNA amounts ↓ Sequencing reactions set up and shipped to sequencing center via overnight delivery ↓ Day 4: DNA sequencing by BRC at Cornell University ↓ Days 5 & 6: DNA sequence analysis and presentation preparation ↓ Day 7: Project Presentation Group Project #4 - Molecular Identification of Bacteria in the Environment Day 1: Day 2: Day 3: Day 4: Days 5 & 6: Day 7: Inoculation of nutrient agar plates ↓ Incubate overnight at 37ºC ↓ Secondary streaking for methylene blue staining and DNA isolation from single bacterial colonies using PrepMan™ Ultra Sample Preparation Reagent ↓ PCR using MicroSeq® 500 16S rDNA PCR kit ↓ Analytical agarose gel of PCR products ↓ Spin purification of PCR products ↓ Analytical agarose gel of purified PCR products and quantification of DNA amounts ↓ Sequencing reactions set up and shipped to sequencing center via overnight delivery ↓ DNA sequencing by BRC at Cornell University ↓ Methylene blue staining of isolates, DNA sequence analysis and presentation preparation ↓ Project Presentation Steps in green same as in human projects Let’s do part of the human bitter taste receptor project right now! A. Phenotyping with phenylthiocarbamide (PTC) taster strips B. Background on the molecular genetics of the PTC taste receptor C. Analyze DNA sequence that “matches” your phenotype - can you find the polymorphisms? D. Location of the amino acid polymorphisms within the 3-D structure of the PTC taste receptor A. What is your PTC taster phenotype? 1. Put paper strip flat on surface of your tongue for a few seconds 2. Repeat with second strip 3. Any difference between the two? 4. Rearrange yourselves into groups based on PTC sensitivity: - strong bitter taste - medium bitter taste - weak bitter taste - no bitter taste B. Phenylthiocarbamide (PTC) taste receptor • The inability to taste certain compounds is usually due to simple, recessive Mendelian inheritance. • Dozens of taste and odorant receptors have been cloned and sequenced in the last 20 years. • The TAS2R28 gene encodes a bitter taste receptor that enables humans to taste the compound PTC. • The PTC (TAS2R28) gene has a single coding exon, for a polypeptide chain with 333 amino acids. B. PTC taste receptor, continued • Three common single nucleotide polymorphisms (SNPs) are associated with PTC sensitivity. • Each SNP results in a change to the amino acid sequence of the PTC receptor. Table 1. Polymorphisms within the PTC gene Position Position SNP Amino Acid (bp) (amino acid) Allele Encoded 145 49 C or G Pro or Ala 785 262 C or T Ala or Val 886 296 G or A Val or Ile • The SNPs are usually inherited together in certain combinations, e.g., haplotypes. Table 2. SNP haplotypes of the PTC gene within two study groups (named for the first letter of the amino acid present at positions 49, 262 and 296) Haplotype European Freq. East Asian Freq. PAV 49% 70% AVI 47% 30% AAV (from recomb. at aa 49) 3% - A later screen identified two additional haplotypes, PVI and AAI, which were found only in individuals of sub-Saharan African ancestry. The AVI haplotype was found in all populations except Southwest Native Americans (Kim et al., 2003). • Certain haplotypes are generally correlated with taster status. Table 3. Genotype association with taste phenotypes (by haplotypes) Genotype (diploid) AVI/AVI (73) AVI/AAV (21) */PAV (170) Nontasters 81% 52% 2% Tasters 19% 48% 98% *= PAV, AVI or AAV. The total number of PTC genotypes observed was 5, as no AAV homozygotes were observed in the study group (Kim et al, 2003). C. Your goal: Examine the DNA sequence chromotograms, locate the areas of the three SNPs and identify ‘your’ amino acid haplotype. Note: remember you are a diploid organism! C. DNA Sequence Analysis of the PTC gene 1. Use sequence handout and Table 1 to determine ‘your’ DNA sequence at the position of each of the three SNPs 2. What is your diploid amino acid haplotype? 3. Does this haplotype correspond with PTC taster status? 4. Any instances of heterozygosity? bp 145 aa #49 C. 4. What about heterozygosity? bp 785 a.a. #262 Note the overlapping blue (C) and red (T) peaks. (The pink N above – inserted when the sequencing computer cannot make a clear call - is a strong hint that something is up at that position!) C. 4. What about heterozygosity, cont.? What are the possible diploid amino acid haplotypes for this individual? Answer: PAV/AVI (most likely) But cannot rule out: PAI/AVV PVV/AAI PVI/AAV (due to recombinant chromosomes in the population) D. Why are people with the AVI/AVI genotype unable to taste PTC? 1. What is the normal structure and function of the PTC receptor? 2. Where within the structure do the non-taster variant amino acids reside? 3. What is the side chain structure of the taster (PAV) and non-taster amino acids (AVI)? 4. How do the variant amino acids alter the structure and/or function of the protein? 1. The PTC receptor is a G-protein linked receptor, with seven transmembrane domains. A taster PTC receptor will bind PTC with its extracellular domain and signal by activating a heterotrimeric G protein on the cytoplasmic surface of the membrane. Pathways of Bitter and Sweet Taste Transduction Margolskee, R. et al., J Biol Chem. 277:1-4, 2002 A non-taster PTC receptor (green above) may either a) not bind PTC b) bind PTC but be defective in activating the heterotrimeric G protein Ligand binding studies indicate that taster and non-taster receptors have the same binding affinity for PTC. Sum: Extracellular domain of receptor likely unaffected in non-tasters. Predicted 3D Structures for PTC Bound to Bitter Receptor Floriano et al. J Mol Model, (2006) 12: 931-941 Taster Variant Non-Taster Variant 2. Location of the Variant Amino Acids in PTC Tasters (PAV) and Non-Tasters (AVI) PTC Tasters (PAV) Non-Tasters (AVI) Amino acid (aa) 49 is located at cytoplasmic base of the first transmembrane region and aa 262 and 296 reside within transmembrane regions #6 and #7, respectively. 3. Amino Acid Structures 49: Pro -> Ala 262: Ala -> Val 296: Val -> Iso 4. Predicted 3D Structure for the PTC Receptor Unbound Bound (note shift in TM6) TM6 Position 262 and TM7 Position 296 Unbound Bound (note change in TM6) For AVI (non-taster) haplotype, V at 262 and I at 296 prevent interaction between transmembrane regions 6 and 7 ‘VI’ mutant variations shown as gray shadows Floriano et al. J Mol Model, (2006) 12: 931-941 D. Conclusions From The Models • The extracellular PTC binding sites between taster and non-taster receptors are not significantly different • Non-tasting in AVI haplotype is likely due to V-262 and I-296; aa 49 has minimal impact • Consistent with PVI = non-taster • AA sites 262 and 296 have the potential to interfere with TM6 and TM7 interaction (V and I are bulkier than A and V, respectively) • It is hypothesized that movement of TM6 is necessary for activation of the heterotrimeric G protein More Fun Informaton about PTC Receptor • PTC taste sensitivity displays a broad and continuous distribution (e.g., it behaves like a quantitative trait). • On average, PTC taste sensitivity is highest for the PAV/PAV (taster) homozygotes, slightly but significantly lower for the PAV heterozygotes, and lowest by far for the AVI/AVI (non-taster) homozygotes. • More rare AVI/AAV heterozygotes have a mean PTC score slightly, but significantly, higher than the AVI/AVI homozygotes. • All non-human primates examined to date are homozygous for the PAV (taster) haplotype. Thus, the AVI nontaster haplotype arose after humans diverged from the most recent common primate ancestor. • There are non-taster chimps: same gene, but different mutation than humans => molecular convergent evolution!!