VA/Duke 4x8 Poster Template
advertisement

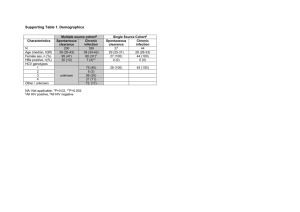
Serum Proteomic Profiles Associated with IL28B Genotype among CHC Patients 1 McCarthy , 1 Cyr , 1 Lucas, 1 Thompson, 2 Patel , Alexander Jeanette J. Derek Joseph J. Will Keyur Hans L. Tillmann,2, Paul J. Clark2, M. Arthur Moseley1 and John G. McHutchison2. 1 2 Thompson , Institute for Genome Sciences and Policy, Duke University, Durham, NC ; 2 Duke Clinical Research Institute and Duke University Medical Center, Durham, NC Background Results • SNP near the gene IL28B (encoding IFN-λ3) are associated with both spontaneous and treatment (Peg-IFN & Riba) induced clearance of hepatitis C virus (HCV). • Subject characteristics: Caucasian (78%); males (61%); mean age 47.4 years; sustained virologic responders to treatment (63%). • The underlying genetic variant(s) responsible for this observed association remains to be identified, yet several SNPs, including rs12979860, appear to be robust markers of its effect. • LC-MS/MS generated 4,186 peptides with positive identifications, which latent factor modeling grouped into 110 metaproteins. • In an effort to gain further insight into the function of this locus, we carried out association analysis of rs12979860 with expression of serum protein quantitative traits in a cohort of patients with chronic hepatitis C (CHC). Methods • Subjects: 41 HCV genotype 1 CHC patients from the Duke Liver Clinic with serum samples available prior to treatment and DNA available for genetic analysis. • IL28B rs12979860 genotyping method: TaqMan. • Two metaproteins significantly associated with IL28B genotype: the liver protein Corticosteroid Binding Globulin (CBG; p = 9.2×10-6) and complement component C5 (p = 0.005). Only CBG remained significant after correction for multiple testing. • rs12979860 ‘C’ (responder) allele was associated with lower levels of CBG metaprotein and explained 38% of the variance in CBG metaprotein. (Figure 1). Figure 1. • CBG metaprotein composition shown in Figure 2. Figure 2. CBG metaprotein peptide composition. Each gray bar is a representation of the labeled protein. Red sections represent polypeptides from isotope groups that are in the factor and black sections represent polypeptides from the isotope groups that are identified in the data set but are not in the factor. Gray represents sections of the protein that are not identified in the data set. % Coverage is the percent of identified peptides from that protein that are in the factor and % signature is the percent of the factor that comes from the associated protein. Note that the sum of % signature is <1 because there are unidentified peptides in the factor that are not shown in the figure. • Proteomic data: generated using LC-MS/MS analysis of pre-treatment serum samples immunodepleted using MARS14 columns and digested with trypsin. • Associations also found between the IL28B rs12979860 genotype and SVR (p = 5.06×10-5) and between CBG metaprotein expression and SVR (p = 0.0011). • Analysis: Peptides with protein identifications were grouped using a latent factor model with informative priors to classify the peptides into 110 metaproteins. Metaproteins were each analyzed for association with IL28B genotype using one-way analysis of variance. • Ingenuity Pathway analysis linked IL28B and CBG via steroid hormones (corticosteroids and progestins) and proinflammatory cytokines (tumor necrosis factor and interferon gamma) (Figure 3) . Figure 3. Ingenuity Pathway Analysis. Conclusions • IL28B rs12979860 is strongly associated with serum levels of corticosteroid binding globulin peptides. • CBG, also known as Transcortin, is a major high-affinity plasma transport protein for glucocorticoids and progestins and is expressed primarily in the liver. • IL28B’s effect on viral clearance may be partly mediated through the action of steroid hormones. • Studies that incorporate direct measures of cortisol and CBG may provide further validation of the role of corticosteroids in IL28B-associated viral clearance. This study was funded in large part by a generous grant from the David H Murdock Institute for Business and Culture via the M.U.R.D.O.C.K. Study and NIH CTSA award 1 UL1 RR024128-01 to Duke University.