in-class version
advertisement

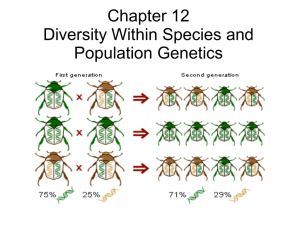
Genetic drift causes allele frequencies to change in populations Alleles are lost more rapidly in small populations Genetic drift results from the influence of chance. When population size is small, chance events more likely to have a strong effect. Sampling error is higher with smaller sample Assume gene pool where frequency A1 = 0.6, A2 = 0.4. Produce 10 zygotes by drawing from pool of alleles. Repeat multiple times to generate distribution of expected allele frequencies in next generation. Fig 6.11 Allele frequencies more likely to change than stay the same. If same experiment repeated but number of zygotes increased to 250 the frequency of A1 settles close to expected 0.6. 6.12c Buri (1956) established 107 Drosophila populations. All founders were heterozygotes for an eye-color gene called brown. Neither allele gives selective advantage. Initial genotype bw75/bw Initial frequency of bw75 = 0.5 Followed populations for 19 generations. Population size kept at 16 individuals. What do we predict will occur in terms of (i) allele fixation and (ii) frequency of heterozygosity? In each population expect one of the two alleles to drift to fixation. Expect heterozygosity to decline in populations as allele fixation approaches. Distribution of frequencies of bw75 allele became increasingly U-shaped over time. By end of experiment, bw75 allele fixed in 28 populations and lost from 30. Fig 6.16 Frequency of heterozygotes declined steadily over course of experiment. Fig 6.17 Effects of genetic drift can be very strong when compounded over many generations. Simulations of drift. Change in allele frequencies over 100 generations. Initial frequencies A1 = 0.6, A2 = 0.4. Simulation run for different population sizes. 6.15A 6.15B 6.15C Populations follow unique paths Genetic drift most strongly affects small populations. Given enough time, even large populations can be affected by drift. Genetic drift leads to fixation or loss of alleles, which increases homozygosity and reduces heterozygosity. 6.15D 6.15E 6.15F Genetic drift produces steady decline in heterozygosity. Frequency of heterozygotes highest at intermediate allele frequencies. As one allele drifts to fixation number of heterozygotes inevitably declines. Alleles are lost at a faster rate in small populations › Alternative allele is fixed Bottlenecks and founder effects are examples of genetic drift. A bottleneck causes genetic drift A bottleneck occurs when a population is reduced to a few individuals and subsequently expands. Many alleles are lost because they do not pass through the bottleneck. As a result, the population has little genetic diversity. A bottleneck can dramatically affect population genetics. Next slide shows effects of a bottleneck on allele frequencies in 10 simulated replicate populations. The northern elephant seal was almost wiped out in the 19th century. Only about 10-20 individuals survived. Now there are more than 100,000 individuals. Two studies in the 1970’s and 1990’s that examined 62 different proteins for evidence of heterozygosity found zero variation. In contrast, southern elephant seals show plenty of variation. More recent work that has used DNA sequencing has shown some variation in northern seals, but still much less than in southern elephant seals. Museum specimens collected before the bottleneck exhibit much more variation than does current population. Clearly, the population was much more genetically diverse before the bottleneck. Founder Effect: when a population is founded by only a few individuals only a subset of alleles will be included and rare alleles may be over-represented. Founder effects cause genetic drift Silvereyes colonized South Island of New Zealand from Tasmania in 1830. Later spread to other islands. http://photogallery.canberra birds.org.au/silvereye.htm 6.13b Analysis of microsatellite DNA from populations shows Founder effect on populations. Progressive decline in allele diversity from one population to the next in sequence of colonizations. Fig 6.13 c Norfolk island Silvereye population has only 60% of allelic diversity of Tasmanian population. Founder effect common in isolated human populations. E.g. Pingelapese people of Eastern Caroline Islands are descendants of 20 survivors of a typhoon and famine that occurred around 1775. One survivor was heterozygous carrier of a recessive loss of function allele of CNGB3 gene. Codes for protein in cone cells of retina. 4 generations after typhoon homozygotes for allele began to be born. Homozygotes have achromotopsia. Achromotopsia rare in most populations (<1 in 20,000 people). Among the 3,000 Pingelapese frequency is 1 in 20. High frequency of allele for achromotopsia not due to a selective advantage, just a result of chance. Founder effect followed by further genetic drift resulted in current high frequency.