Section F
advertisement
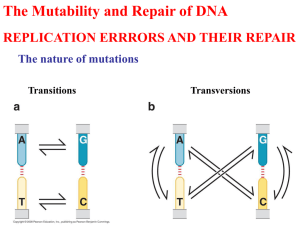
Section F DNA Damage, Repair and Recombination F1 Mutagenesis F2 DNA Damage F3 DNA Repair F4 Recombination Section F: DNA Damage Repair Yang Xu, College of Life Sciences F1 Mutagenesis • • • • Mutation Chemical mutagens Direct mutagenesis Indirect mutagenesis Section F: DNA Damage Repair Yang Xu, College of Life Sciences Mutation-I • Definition: Mutations are permanent, heritable alteration in the base sequence of DNA. • Classification: 1. Point mutation: A single base change, includes: transition: one purine (or pyrimidine) is replaced by the other purine (or pyrimidine); transversion: a purine replaces a pyrimidine. 2. Frame-shift mutation : Insertions or deletions involve the addition or loss of one or more bases,and the translated protein sequence to the C-terminal side of the mutation is completely changed. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Mutation-II Phenotypic effects of mutation: • Silent mutation: If the mutation is (1) in a non-coding or nonregulatory piece of DNA; (2) in the third position of a codon. • Missense mutation: If the mutation results in an altered amino acid in a gene product, then it is a missense mutation whose effect can vary from nothing to death, depending on the position of the amino acid affected. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Mutation-III Consequences of mutation: • Tumorigenesis: Mutations that affect the processes of cell growth and cell death can result in tumorigenesis; • Genetic polymorphisms: The accumulation of many silent and other non-death mutations in populations produces genetic polymorphisms. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Chemical mutagens Most chemical mutagens are carcinogens • Base analogs: are the normal bases with altered base pairing properties and can cause direct mutagenesis. • Nitrous acid: deaminates cytosine to produce uracil, which base-pairs with adenine and causes G•CA•T. NH2 N 0 O HNO2 N C HN De-amination 0 N U • Alkylating agents: produce alkylated (methylated) bases. • Intercalators: generate insertion and deletion mutation. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Direct mutagenesis • Mechanism: If a base analog is not removed by a DNA repair mechanism before passage of replication fork, then an incorrect base will be incorporated. The second replication fixes the mutation permanently in the DNA. • For example: thymine analog 5-bromo-uracil (B) base-pairs with adenine (A) and also with guanine (G). After one round of replication, thus the B causes A•T G•C. AGCTTCCTA TCGAAGGAT TB AGCTBCCTA TCGAAGGAT Section F: DNA Damage Repair AGCTBCCTA TCGAGGGAT AGCTBCCTA TCGAAGGAT B:A BG AGCTCCCTA TCGAGGGAT AGCTBCCTA TCGAAGGAT B:A GC Yang Xu, College of Life Sciences Indirect mutagenesis • Definition: Indirect mutagenesis results from: 1. The activation of trans-lesion DNA synthesis (SOS) 2. The suppression of proofreading system, one or more incorrect bases are incorporated in DNA. • Mechanism: 1. Most lesions in DNA are repaired by error-free direct reversal or excision repair mechanisms before passage of a replication fork. 2. If this is not possible, trans-lesion DNA synthesis may take place and one or more incorrect bases become incorporated opposite the lesion, so as to maintain the integrity of the chromosome. 3. Proofreading may be suppressed during this process. Section F: DNA Damage Repair Yang Xu, College of Life Sciences F2 DNA Damage • • • • • DNA lesions Spontaneous lesions Oxidative damage Alkylation Bulky adducts Section F: DNA Damage Repair Yang Xu, College of Life Sciences • Definition: A lesion is an alteration of the normal chemical or physical structure of the DNA. • Mechanism: Some of the nitrogen and carbon atoms in the heterocyclic ring systems are chemically quite reactive. Many exogenous agents, such as chemicals and radiation, can cause structure changes to these positions and result in DNA lesions. • Consequences: 1. DNA mutation: If such a lesion was allowed to remain in the DNA, a mutation could become fixed in the DNA by direct or indirect mutagenesis; 2. Cell death: If the chemical change may produce a physical distortion in the DNA which blocks replication and/or transcription, causing cell death. Section F: DNA Damage Repair T A … DNA lesions C G –A:T – G C H C OH G T A T A T A C G A:T…... DPC G:C…... T:A Yang Xu, College of Life Sciences Spontaneous lesions • Mechanism: 1. Deamination: Cytosine deaminates to produce uracil. Any uracil found in DNA is removed by an enzyme called uracil DNA glycosylase and is replaced by cytosine. 2. Depurination: It is another spontaneous hydrolytic reaction that involves cleavage of the N-glycosylic bond between N-9 of the purine bases A and G and C-1' of the deoxyribose sugar and hence loss of purine bases from the DNA backbone. 3. Depyrimidination: it can also occur with very low frequency. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Oxidative damage • Definition: Reactive oxygen species (ROS) such as super-oxide and hydroxyl radicals produce a variety of lesions, such damage occurs spontaneously but is increased by some exogenous agents including -rays. • Examples: NH2 -CH2OH 8 =OH H2 N 8-Oxoguanine Section F: DNA Damage Repair HO 2-Oxoadenine O 5-hydroxymethyluracil Yang Xu, College of Life Sciences Alkylation • Examples for alkylating agents: O CH3–S–O–CH3 O H2N–C–N O • CH2CH3 N=O Examples for methylated bases: CH3 NH2 H2N CH3 Section F: DNA Damage Repair CH3 H2 N Yang Xu, College of Life Sciences Bulky adducts • Definition: They are bulky covalent adducts which are formed by physical or chemical agents. They result in loss of base pairing ability with the opposite strand. • Examples: 1. Physical agent: UV light. C–N S-N C=O 2. Chemical agent: Aromatic arylating C=C agents P C–N G–NH S-N C=O C=C P450 Section F: DNA Damage Repair Yang Xu, College of Life Sciences F3 DNA Repair • Photo-reactivation • Alkyltransferase • Excision repair • Mismatch repair Section F: DNA Damage Repair Yang Xu, College of Life Sciences Photo-reactivation • Definition: In the presence of visible light, the DNA photolyases (photo-reactivating enzymes) can resolve cyclo-butane pyrimidine dimers into monomer again, this process is known as photo-reactivation. • Mechanism: These enzymes have prosthetic groups which absorb blue light and transfer the energy to the cyclo-butane ring which is then cleaved. Photoreactivation is specific for pyrimidine dimers. It is a direct reversal of a lesion and is error-free. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Alkyltransferase Mechanism: Mutagenic protection is afforded by an alkyltransferase, which removes the alkyl group from the directly mutagenic O6alkylguanine. The alkyl group is transferred to the protein itself and inactivates it. Thus, each alkyltransferase can only be used once. Cys-SH Cys-SHCH3 CH3 H2 N H2 N • Photo-reactivation and alkyl-transferase effect are all belong to the error-free direct reversal forms. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Nucleotide excision repair (NER) • Nucleotide excision repair (NER): In NER, an endonuclease makes nicks on either side of the lesion, then the lesion is removed to leave a gap. This gap is then filled by a DNA polymerase I, and DNA ligase makes final phosphodiester bond. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Base excision repair (BER) • Base excision repair (BER): the lesion is removed by a specific DNA glycosylase, which cleave the N-glycosylic bond between the altered base and the sugar. This results AP site, then it is cleaved and expanded to a gap by an AP endonuclease plus exonuclease. This gap is then filled by a DNA polymerase I, and DNA ligase makes final phosphodi-ester bond. Section F: DNA Damage Repair Yang Xu, College of Life Sciences Mismatch repair • Mechanism: 1. Mismatch: In a replicational mispair, for example G•T or C•A, the wrong base is only in the daughter strand. For mismatch repair, wrong pair and wrong strand must be known. 2. Distinguish: the wrong pair (mismatch) is identified and attached by MutS/MutL protein complex, and the wrong (daughter) strand is identified by MutH endonuclease according to the hemi-methylated sequence GATC. 3. Excision repair: MutH endonuclease then makes nicks at GATC site on either side of the lesion in the newly synthesized strand, and do excision repair. Section F: DNA Damage Repair Yang Xu, College of Life Sciences F4 Recombination • Homologous recombination • Site-specific recombination • Transposition Section F: DNA Damage Repair Yang Xu, College of Life Sciences Homologous recombination-I • Definition: This process involves the exchange of homologous regions between two DNA molecules, it is also known as postreplication repair. The integrity of DNA containing un-repaired lesions can be fixed during replication by homologous recombination. • Mechanism: 1. In eukaryotes, this commonly occurs during meiosis, when the homologous duplicated chromosomes line up in parallel in metaphase I and the non-sister chromatids exchange equivalent sections by crossing over. From father From mother Section F: DNA Damage Repair Yang Xu, College of Life Sciences Holliday model 2. In prokaryotes: This model was proposed by Robin Holliday in 1964. Mechanism: Two homologous DNA align with each other; Nuclease/RecBCD protein complex makes nicks near the chi sequence (GCTGGTGG); The ssDNA becomes coated in RecA protein to form RecA-ssDNA filaments. They cross over and search the opposite sequence; The nicks are sealed and a four-branched Holliday structure is formed, then branch migrates; The Holliday intermediate can be resolved into two DNA duplexes in one of two ways: Section F: DNA Damage Repair A B a A b B a A b B a b Yang Xu, College of Life Sciences Holliday model Section F: DNA Damage Repair A B a A b B a A b B a b Yang Xu, College of Life Sciences Site-specific recombination • Definition: is mediated by proteins that recognize specific DNA sequences. It does not require RecA or ssDNA. • Examples: Prokaryotes: Bacteriophage DNA / E. coli DNA attP of λ p O p’ B B’ attB of E.coli Int (Integrase) B Section F: DNA Damage Repair O P’ O Xis (Excisionase) P O B’ Yang Xu, College of Life Sciences Transposition Definition: Transposons are small DNA sequences that can move to almost any position in a cell’s genome. Transposition has also been called illegitimate recombination because it requires no homology between sequences nor is it site-specific. Examples: IS in E. coli Insertion Sequence: IS • They comprise a transposase gene flanked by ITR. • The transposase makes a staggered cut in the chromosomal DNA and then inserts a copy of IS elements. • The gaps sealed by DNA polymerase I and DNA ligase, resulting in a duplication of the target site. • The genes between two copies of a transposon can be deleted by recombination between them. Section F: DNA Damage Repair transposase --CGTAAGCTGCGGCAGGATTGA-transposase --GCATTCGACGCCGTCCTAACT---CGTAAGCT --CGTAAGCTCGCCG GCGGCAGGATTGA-- --GCATTCGACGCCG TCCTAACT-CGCCGTCCTAACT-- --CGTAAGCTCGCCG --CGTAAGCTCGCCGGCGGCAGGATTGA-GCGGCAGGATTGA---GCATTCGACGCCG --GCATTCGACGCCGCGCCGTCCTAACT-CGCCGTCCTAACT-Yang Xu, College of Life Sciences Transposition 1983. Barbara McClintock (86y) DNA transposable elememt Section F: DNA Damage Repair Yang Xu, College of Life Sciences That’s all for Section F Section F: DNA Damage Repair Yang Xu, College of Life Sciences