Class11
advertisement

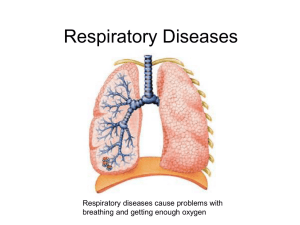
Stem Cells and Lineages: Mechanisms of Lung Development • • • • • • • Overview of stages of lung development Epitheial-Mesenchymal interactions Budding Branching morphogenesis Sacularization and Alveolarization (Maturation) Genes and growth factors that control these processes Focus on how these molecules were shown to play a role in the processes Stem Cells and Lineages: Mechanisms of Lung Development • • • Lungs did not develop from gills, they are a completely separate structrure Developed from swim bladders in fish Gills develop from pharyngeal arches independently of lungs. Bladder Early Late Review of mouse lunglung development Overview of mouse development Canalicular Pseudoglandul ar Saccular Alveolar Cordoso, Dev. Dyn. 219:121 (2000) Neonatology: SE Wert 5th edition 1999. Overview of mouse development Review of mouse lung lung development Canalicular Pseudoglandul ar Saccular Alveolar Cordoso, Dev. Dyn. 219:121 (2000) Neonatology: SE Wert 5th edition 1999. Summary of stages Nodal (TGF member) Foxa2, Sox17, GATA4,6, Stat3 needed for gut tube formation as shown by KOs Bladder Early Late Budding Evidence for players 1) Tissue recombination experiments (70 years ago to today) 2) In situ hybridization (guilt by assocation) 3) Gene KOs and transgenics 4) In vitro culture assays with beads and factors 1) Tissue recombination experiments (70 years ago to today) Rudnick, D. 1933 Chick lung rudiments transplanted onto Chorioallantoic membranes and cultured. Mesenchyme required for branching. Wessells, N 1970 Lung buds form opposite sites where lung mesenchyme is placed in vitro. Masters, JRW 1976 Mesenchyme is needed to induce branching morphogenesis in vitro from tracheal buds. Shannon, JM 1994 Distal mesenchyme can induce bud and branching of proximal trachea Shannon, JM 1994 Distal mesenchyme can induce bud and branching of proximal trachea Shannon, JM 1994 Distal mesenchyme can induce bud and branching of proximal trachea Day 13 lung Grafted Trachea time 0 Grafted Trachea 24hrs Shannon, JM 1994 Distal mesenchyme can induce bud and branching of proximal trachea Shannon, JM 1994 Distal mesenchyme can induce bud and branching of proximal trachea Get relatively normal looking lung lobes when mesenchyme is grafted next to trachea. What are inducing molecules from normal E99.5 gut mesenchyme that induce budding? FGF10 is believed to be the primary bud inducing molecule. Retinoic acid receptors are also implicated in initial lung formation as double KOs of 2 prevent trachea formation. Evidence for FGF10? 1) Is expressed in mesenchyme opposite where lung bud forms. 2) KO of FGF10 prevents budding and lung formation. Little else known about bud formation How is FGF10 localized? What target genes are there in the epithelium that respond to FGF10? Branching morphogenesis • FGF10, TTF-1, Shh other molecules in a repeating signalling network during branching • Evidence primarily from KOs and explant studies Branching morphogenesis Foxa1 or Foxa2 needed for Shh expression in epithelium FGF9-Shh network also implicated in early branching. KO reduces branching. Development 133, 1507-1517 (2006) doi:10.1242/dev.02313 Branching lineage Genetic control of branching lineage Spry2 (Sprouty 2) a member of a tyrosine kinase inhibitor family. Ectopic domain branching in Spry22/2 mutants. a, The RCd lobe (ventral view) of an E12.5 control (Spry21/2) lung with a single secondary branch (V1, circled) off RCd, at the level of RCd.L4 (L4). Below, RCd.V1 lineage and schematic of RCd with a single ventral secondary branch (V1). b, Same view of an E12.5 Spry22/2 lung showing the normal ventral secondary branch (V1) and an ectopic branch (V*) that forms earlier and proximal to V1. V* has already sprouted additional generations of branches. Below, lineage and schematic show V* plus additional ectopic ventral branches (V**, V***; dashed lines in schematic) seen in other Spry22/2 lungs (Supplementary Fig.4). Scale bar (for a and b), 200 mm. Genetic control of branching • Shh (Epith.), Hip1, Ptc1,Gli2, Gli3 (mesen.) • FGF10, FGF9 (mesen.) • TTF-1 aka Titf1 (Epith.) • Foxa1+Foxa2, single KOs branch normally req. for Shh expression in epith. • BMP4 (Tgf family) (mesen.) • Pod1 (bHLH TF in mesen.) • -catenin signalling in epith. And mesen. • Foxf1 (mesen.) • Tbx4, 5 req. for Fgf10 express. in mesen. (siRNA studies) • Hoxa5 (mesen.) • Spry2 FGF tyrosine kinase inhibitor (Epith.) Genetic control of branching Reciprocal Epithelial-Mesenchymal interactions mediated by growth factor- and cell contact-regulated transcriptional networks. These interactions both constitute a program of growth, morphogenesis and differentiation and then help establish the quazi-stable epigenetic states that characterize each differentiated cell. Overview of mouse development Review of mouse lung lung development Canalicular Pseudoglandul ar Saccular Alveolar Cordoso, Dev. Dyn. 219:121 (2000) Neonatology: SE Wert 5th edition 1999. Canalicular and saccular phases (later maturation E16.5-P5) • Transition from undifferentiated to fully differentiated cells and marked reduction in cell proliferation. • KOs, conditional KOs and conditional transgenics • In vitro culturing of lungs Canalicular phase (E16.5-17.5) what cell types are formed in conducting airways? Foxj1 KO eliminates cilia on cells p63 KO eliminates basal cells Gfi-1 KO reduces number of PNECs Early Saccular phase (E17.5-E18.5) Hypoplastic versus hyperplastic lungs • Coupling of redued cell proliferation to increased cell differentiation • Some mutants have smaller lungs that are "immature", other mutants have larger lungs that are "immature" • Maturation measured by terminal differentiation markers in Type II and Type I epithelial cells. Saccular phase (E17.5-P5) Nfib (hyperplastic) Lung defects in Nfib-/- and +/- E17.5 animals -/- E17.5 lungs look like E15.5 lungs suggests maturation defect Increased DNA content in E18.5 Nfib-/- lungs # of Genotype pups Nfib +/+ 6 Nfib -/+ 12 Nfib -/5 Total body weight (g) 1.18 0.065 1.21 0.124 1.38 0.083 Left lung DNA/ Total body wt. (ug/g) 38.6 3.93 51.9 5.08 73.1 10.47 ~4X increased PCNA levels in E18.5 Nfib-/- lungs (RT-QPCR) Loss of differentiation markers reflect immaturity in Nfib-/- lungs (RT-QPCR) E15.5 E17.5 gene Nfib+/+ Nfib-/Nfib+/+ Nfib-/+ Nfib-/Sftpa 2** 2 100* 40 10 Sftpb 6 3 100 85 39 Sftpc ND ND 100 74 48 Shh 146 215 100 ND 214 Vegfa188 20 37 100 39 17 *Signal from E17.5 wild type animals was set to 100% for each gene and data is reported relative to E17.5 WT controls. (ND, not determined) Standard deviation 10-15%. Saccular phase (E17.5-P5) TGFb3 KO (hypoplastic) E18.5 +/+ -/- +/+ -/- Saccular phase (E17.5-P5) TGFb3 KO (hypoplastic) E18.5 loss of diffn. markers -/- Early Saccular phase (E17.5-E18.5) Hypoplastic versus hyperplastic lungs • Coupling of reduced cell proliferation to increased cell differentiation can be affected differently in different mutants. • Maturation measured by terminal differentiation markers in Type II and Type I epithelial cells. Some mutations affect expression or proliferation of both cells types, some only one. • How many maturation programs are there and how are they linked? Saccular phase (E17.5-P5) Control of epithelial differentiation I Saccular phase (E17.5-P5) Control of epithelial differentiation II This leaves out NFI, GR, Gata6, SP3, Sox11, HIF2A, TGF3 and other TFs and GFs that affect maturation. Nfib Alveolarization (P5-P28) • Terminal differentiation of type I cells • Further reduction in mesenchyme • Increased surfactant expression in type II cells • Expansion of saccules into alveoli with septation Just so you don't think it's too simple Just so you don't think it's too simple Just so you don't think it's too simple Just so you don't think it's too simple Things we left out Vasculature Repair of damage Lung Stem Cells Lung diseases Immune responses Review of mouse lunglung development Overview of mouse development Canalicular Pseudoglandul ar Saccular Alveolar Cordoso, Dev. Dyn. 219:121 (2000) Neonatology: SE Wert 5th edition 1999. Overview of mouse development Review of mouse lung lung development Canalicular Pseudoglandul ar Saccular Alveolar Cordoso, Dev. Dyn. 219:121 (2000) Neonatology: SE Wert 5th edition 1999. How to build an organ Reciprocal Epithelial-Mesenchymal interactions mediated by growth factor- and cell contactregulated transcriptional networks. These interactions both constitute a program for growth, morphogenesis and differentiation and then help establish the quazi-stable epigenetic states that characterize each differentiated cell. Many organs follow this strategy with the addition of other processes such as cell-recruitment, Epithelial-Mesenchymal transitions (EMT) and Mesenchymal-Epithelial transitions (MET).