The plant of the day
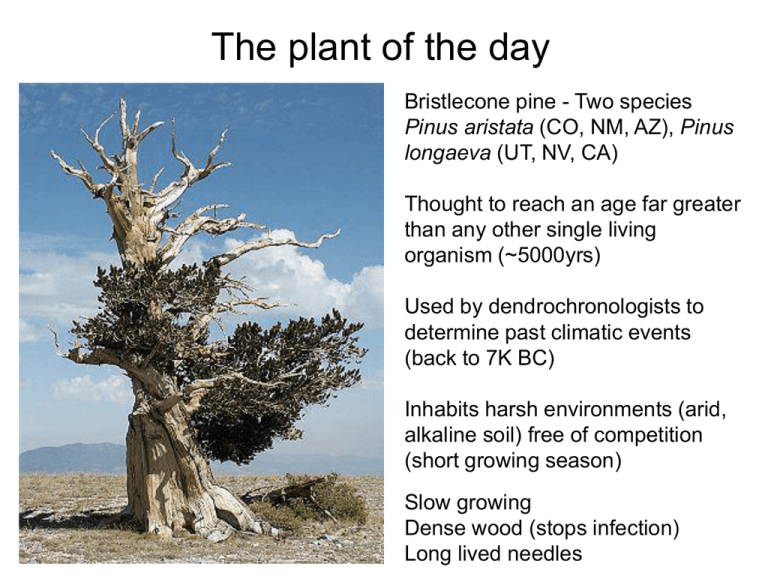
Bristlecone pine - Two species
Pinus aristata (CO, NM, AZ), Pinus
longaeva (UT, NV, CA)
Thought to reach an age far greater
than any other single living
organism (~5000yrs)
Used by dendrochronologists to
determine past climatic events
(back to 7K BC)
Inhabits harsh environments (arid,
alkaline soil) free of competition
(short growing season)
Slow growing
Dense wood (stops infection)
Long lived needles
Non-random mating, genetic drift, and
population structure
Non-random mating
Assortative mating – mating
with individuals that are similar
or dissimilar for a given trait.
Inbreeding – mating with a close
relative.
Positive Assortative Mating
If the phenotype is under genetic control, Positive
assortative mating increases homozygosity and
decreases heterozygosity for the genes affecting the trait.
Positive
Assortative Mating
in the genus Burmeistera, bats
are more efficient at moving
pollen between wide flowers,
whereas hummingbirds excel at
pollen transfer between narrow
flowers.
Negative Assortative Mating
If the phenotype is under genetic control, Negative
assortative mating increases heterozygosity and
decreases homozygosity for the genes affecting the trait.
Negative Assortative Mating
Plant self-incompatibility
systems lead to negative
assortative mating.
Examples: Sunflowers
Cocoa tree
Blue bells
Brassica rapa
(field mustard)
Inbreeding
Inbreeding: mating with a close relative
Biparental: two different individuals are involved
Extreme inbreeding
Intragametophytic selfing:
mating between gametes
produced from the same
haploid individual
-100% homozygosity in
one generation!
- some ferns and mosses
The effects of inbreeding on genotype
and allele frequencies
Fewer heterozygotes and more homozygotes
No change in allele frequency
Inbreeding
Inbreeding does NOT change allele frequency by itself
It does increase homozygosity
Inbreeding coefficient (F):
measures the extent to which populations depart from the
expectation of 2pq (remember p² + 2pq + q² = 1)
He = Expected heterozygosity, HW (2pq)
Ho = Observed heterozygosity
F = (He-Ho)/He
Evolutionary Consequences of
Inbreeding
In large, random
mating populations,
most individuals will
not suffer from
deleterious effects of
recessive deleterious
alleles
Under inbreeding,
increased
homozygosity for these
recessive deleterious
alleles results in
reduced fitness
Genetic drift
Definition: Changes in allele frequency due to random sampling.
One of the requirements for the maintenance of stable allele
frequencies in populations is a very large population size.
Genetic drift is the consequence of finite population size.
Genetic drift
Alleles that do not affect
fitness fluctuate randomly in
frequency, which eventually
results in the loss of alleles
from populations. One allele
becomes fixed.
Genetic drift
Different populations will lose different alleles. The probability
that a particular allele will be fixed in a population in the future
equals the frequency of the allele in the population.
If a large
number of
populations is
considered,
each drifting,
the total
heterozygosity
overall will
decrease.
Genetic drift
Starting with a
population size of N with
two alleles in equal
frequencies p and q, the
likely magnitude of
divergence from the
initial frequencies
increases with time.
Genetic drift
After 2N generations, all
allele frequencies are
equally likely.
The average time to
fixation of one of the
alleles is 4N generations.
Effective population size
Effective population size - number of individuals in the
population that successfully pass genes to the next generation.
-usually smaller than the actual population (census) size
-drift will occur more quickly in smaller populations
Effective population size and Drift
Effective population size
The effective population size (Ne) is affected by biological
parameters other than the number of breeding individuals in the
population. These include:
•Variation in offspring number
among individuals
•A sex ratio other than 1:1
•Natural selection
•Inbreeding (reduces the number
of different copies of a gene
passed to the next generation)
•Fluctuations in population size
Founder effects
When a small number of individuals from a source population establish a new
population genetic variation can be lost. The loss of genetic variation due to
such an extreme bottleneck is called the founder effect.
Simulations of founder effects
suggest that a small number
founders and a small population
growth rate (r) result in greater loss
of genetic diversity.
Eventually mutation will restore
genetic variation in a founding
population.
Effects
ofsummary
Drift
Genetic
Drift:
• Within populations
– Changes allele frequencies
– Reduces variance
– Does not cause deviations from HW expectations
• Among populations (if there are many)
– Does NOT change allele frequencies
– Does NOT degrade diversity
– Causes a deficiency of heterozygotes compared
to Hardy-Weinberg expectations (if the existence
of populations is ignored), like inbreeding.
Effects
ofisDrift
Genetic
drift: why
it important?
• Erodes genetic variation within populations
• Causes population differentiation
• Strength is dependant on population size
• The demographic history of populations effects patterns of
genetic variation
• Can oppose selection- conservation implications
• Provides a “neutral” model for evolutionary change and most
molecular changes are effectively neutral
Population structure
How do we measure population genetic structure?
Sewall Wright
Wright’s fixation index
Fixation index is a measure of genetic differentiation among
populations
Compare heterozygosity at different hierarchical levels
FST=(HT-HS)/HT
HT: The overall expected HW heterozygosity for the total area
HS: The average expected HW heterozygosity among organisms
within populations
Linanthus parryae population
structure
What is the genetic
divergence among sub
populations FST?
What could be causing
the divergence in flower
colour among the sub
populations?