ForwardGeneticsMapping2012
advertisement
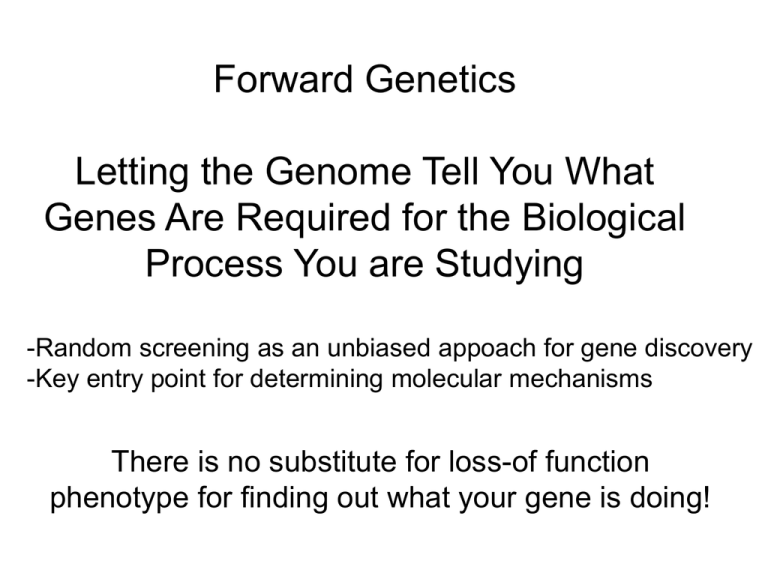
Forward Genetics Letting the Genome Tell You What Genes Are Required for the Biological Process You are Studying -Random screening as an unbiased appoach for gene discovery -Key entry point for determining molecular mechanisms There is no substitute for loss-of function phenotype for finding out what your gene is doing! A Little Genetics Genotype: what alleles you have (heterozygous, homozygous, etc.) Phenotype: what you look like (wild type or mutant for some trait) Recessive allele: genotype needs to be homozygous mutant in order to see mutant phenotype Dominant allele: have mutant phenotype even when heterozygous for mutant allele Zygotic gene: genotype of embryo determines phenotype of embryo Maternal effect gene: genotype of MOTHER determines phenotype of embryo A-P polarity set up in egg chamber bicoid mRNA microtubule-based - oskar mRNA protein + nanos RNA + bicoid, oskar and nanos are examples of genes acting in the OVARY that influence patterning of the EMBRYO Therefore, the GENOTYPE of the MOTHER determines the PHENOTYPE of the EMBRYO (Maternal Effect) Examples: Zygotic recessive (phenotype) m/+ X m/+: m/+ (wild type) +/+ (wild type) m/m (mutant) Zygotic dominant m/+ X m/+: m/+ (mutant) +/+ (wild type) m/m (mutant) Maternal effect recessive Mom Dad Embryo (phenotype) m/+ X m/+: m/+ (wild type) +/+ (wild type) m/m (wild type) Mom Dad m/m X m/+: Embryo m/+ (mutant) m/m (mutant) Mom m/+ X Embryo m/+ (wild type) m/m (wild type) Dad m/m: Types of Mutants Hypomorph: Loss of function Often recessive - genotype needs to be homozygous for mutation (m/m) to see phenotype Amorph or Null: Complete loss of function (behaves like a deletion of gene) Hypermorph: Gain of function Often dominant - can see a phenotype even if genotype is only heterozygous (m/+) Antimorph: Behaves stronger than null (e.g. dominant negative) Neomorph: New function (e.g. gene now expressed in ectopic location) Chemical Mutagenesis EMS (Ethyl methane sulfonate): e.g. flies ENU (n-Ethyl-n-nitrosourea): e.g fish and mice Chemically modify DNA bases to induce replication errors Preferentially induce point mutations (but some small deletions) Advantages: Most random of mutagens Alleles of different strengths Disadvantages: Harder to identify lesion (clone gene) Radiation Mutagenesis X-rays and gamma rays most common Induce double strand DNA breaks -Deletions -Inversions -Translocations Advantages: Often produce null mutations Easy to identify lesion (often just by looking at chromosomes) Disadvantages: Usually take out multiple genes Insertional Mutagenesis (Transposon or Retroviral) Insertion of transposon or viral sequences affects gene function Can control transposon jumping by separating transposase from transposon Advantages: Easy to identify lesion, clone gene (gene is “transposon tagged”) Disadvantages: Non-random integration/mutagenesis -affects target distribution -affects allele strength Currently being done in flies, fish and mouse Traditional Screens for Recessive Mutations EMS Make mutant sperm Make heterozygous mutant individuals Make mutant brothers and sisters Cross heterozygous brothers and sisters to make homozygous mutant offsprint Nusslein-Volhard and Weischaus Nobel Lectures http://www.nobel.se/medicine/laureates/1995/ Summary of X, 2 and 3 Nusslein-Volhard and Weischaus Nobel Lectures http://www.nobel.se/medicine/laureates/1995/ paired WT knirps Large Scale Forward Mutagenesis in Zebrafish Nusslein-Volhard and Dreiver Labs 1996 Problems with traditional homozygous mutant screens: -Genes are not all equally mutable -some are small targets or tough to induce loss of function (e.g. microRNAs) -Genes can be redundant -knocking out one copy doesn’t always give phenotype -Genes are pleiotropic -if embryo dies before your process “happens”, can’t tell if that gene is required Sensitized Genetic Screens: the sevenlessTS screen Simon, Bowtell, Dodson, Laverty and Rubin, 1991 sevenless: receptor tyrosine kinase required for R7 specification Problem: sev is specific to eye, but downstream RTK pathway common to all RTKs and therefore embryonic lethal How do you identify sev pathway components (and therefore components of all RTK signaling)? Sensitized Genetic Screens: the sevenlessTS screen Simon, Bowtell, Dodson, Laverty and Rubin, 1991 sevTS: Making flies “on the edge” 22.7°C R7 mostly present--screen for dominant enhancers of sev -R7 now lost 24.3°C R7 mostly absent--screen for dominant suppressors of sev -R7 now restored e.g. ras ras-/ras+ = wild type ras-/ras- = uninformative dead embryo sevTS 24.3°C with ras-/+ = no R7 cell Normally ras is recessive but now behaves as a dominant enhancer of sevTS Sensitized Genetic Screens: generic eye screens Express programmed cell death gene in eye Look for suppressors/enhancers of programmed cell death pathway Mosaic Screens Genetic mosaics can be created by mitotic recombination induced by X-rays or a site-specific recombinase FLP/FRT in Drosophila: Golic and Lindquist, 1989 Essential reagents: Xu and Rubin, Chou and Perrimon Treisman Lab Forward/Reverse Genetics: Whole genome RNAi Screens Advantages: -Save gene identification step Disadvantages: -Depends on genome sequence/annotation -Efficiency of RNAi is variable -Issues with delivery of dsRNA trigger Cell Culture: Transfect or bathe cells with dsRNA Cell Culture: Lentiviral vectors expressing dsRNA C. elegans: Feed worms bacteria expressing dsRNA Flies: Inject embryos with dsRNA Flies: UAS-shmiRNAs for every gene in genome Positional Cloning Finding a needle in a haystack Finding a single bp change in 3.6 x 108 (Fly) 3.4 x 109 (Fish) 6.0 x 109 (Human/Mouse) First fly gene: Ultrabithorax 1979 First human disease genes: chronic granulomatous disease 1986 Duchenne muscular dystrophy 1987 Cystic fibrosis 1989 (approx. 1200 disease genes now cloned) First fish gene: one-eyed pinhead 1998 How do you find your gene? Identify a transposon-induced allele of your gene: transposon then “tags” genomic region of interest Find a genomic lesion (deletion/inversion/transposition) allele of your gene: breakpoints in genome identify region of interest For point mutants: meiotic mapping and positional cloning Genetics 101 Mendelian Inheritance Linkage a ;b + ;+ a b X + + a b a ;b ; X + + a b a ; b + ;+ + ; b a ;+ a b a b a b a b 1 : 1 : 1 : 1 Parentals = Recombinants a b a b X + + + + a b + + X a b a b + b a + + + a b a b a b Parentals > Recombinants 160 40 % Recomb Recombination = # Recombinants X 100 = Map Units Frequency Total CentiMorgans (cM) 40/200 x 100 = 20 cM a b a b Three genes: a, b, c a c a c a c + + Meiotic Mapping X + + + + c b c b X + + + + X a c a c c b + + X c b c b + c a + + + a c a c a c Recombinants Parentals 190 10 10/200 x 100 = 5 cM a c a c a + + c b Parentals c b c b c + c b Recombinants 170 30 30/200 x 100 = 15 cM c 5 + b c b b 15 To Clone Gene C a 1) 2) 3) 4) c b Link meiotic map to physical map (DNA) Identify markers and map crossovers to define limits of C Identify genes within this region Determine which gene is C Markers for Meiotic/Physical Mapping “Classically” done using visible dominant and recessive mutations -Low density of useful markers -Less rooted in physical map Can improve the density of visible markers using transgenes e.g. w+ transposons in flies Modern methods directly assess DNA polymorphisms Random markers e.g. Randomly Amplified Polymorphic DNA (RAPDs) PCR w/ primers of random sequence, get few random products Presence or absence of product can depend on as little as single bp change Don’t require prior knowledge of genome sequence Allows “entry” into physical map (identifies STS near gene of interest) Simple sequence length polymorphisms (microsatellite DNA, e.g. CA repeats) PCR shows small polymorphic changes in repeat number Advantage: easy to analyze Disadvantage: Not enough (low density) Single nucleotide polymorphisms (SNPs) Advantage: Maximum possible density Disadvantage: Can be difficult to assay Syvanen, Nat Rev Gen 2001 SNPs alter oligo annealing Suitable for microarrays SNPs alter oligo annealing +/- PCR product Single nucleotide “mini-sequencing” Suitable for microarrays Afymetrix offers SNP Chips that can genotype 10-50,000 SNPs Also, -Single strand conformation polymorphisms (detected in gels) -Denaturation HPLC -Mass-spec DNA sequencing Sounds easy but… -Compare mutagenized chromosome with interesting phenotype to control, parental chromosome that was isogenized before screen -Found 165 sequence changes on third chromosome. -Could only verify 103 (some false positives). Others likely not found (false negatives) since not all regions have good sequence. -Of these, 11 made changes to ORFs. Therefore, still some work to figure out correct gene. The First Association Between the Meiotic and Physical Maps RAPDs Allele 1: products A and B A B Allele 2: Change in site 2 No product A http://avery.rutgers.edu/WSSP/StudentScholars/project/archives/onions/rapd.html Bulk Segregant Analysis: Look for Linkage Start with mutation heterozygous in strain 1 Use strain 2 as polymorphic mapping strain Strain 1: m/+ a1 m b1 a1 + (lack of recombination) Strain 2: +/+ X b1 a2 + a2 + a1 m a2 + X b2 b2 b1 b2 Mutant = m Polymorphic Markers = a1, a2, b1, b2 Mutant ‘bros X Backcross to Parental Strain 1 a1 a1 m + wt ‘bros a1 m a1 a1 m m a1 m a2 + a1 m a2 + a1 m b1 b1 b2 b1 b2 b1 b1 b1 b1 b1 Sort mutant vs. wt ‘bros Make DNA from pools a1 + b1 Any combo b is mixed 1 and 2, therefore unlinked to m a is always (or mostly) 1, therefore linked to m (few recombinations) 1) Identify closely linked polymorphic markers oep -Bulk segregant analysis identifies 15AH and 20K as “close” to oep Now look for recombinants between closely linked marker and gene Backcross to Strain 1: m/+ a1 m a1 + b1 b1 Strain 2: +/+ X a2 + a2 + a1 a2 m X + b2 b2 b1 Mutant = m Markers (alleles) = a1, a2, b1, b2 a2 m b2 a1 m b1 b2 X Parental Strain 1 a1 a1 m + b1 b1 Examine INDIVIDUAL offspring = individual meioses (gametes) of parents -Look for recombination between close marker and gene -Screen 3100 “meioses” to find rare recombinants -Save Recombinants: can go back and analyze later with new markers to further define WHERE recombination took place and therefore limits of WHERE oep can be! 1) Identify closely linked polymorphic markers oep -Bulk segregant analysis identifies 15AH and 20K as “close” to oep -Analyze DNA from 3122 INDIVIDUAL mutant ‘bros -find 1 recombinant b/w 15AH and oep 0.03 cM = 18 kb (IF 600 kb/cM) -find 5 recombinants b/w 20K and oep 0.16 cM = 96 kb Now close enough to go after DNA in region (link meiotic map to physical map) 2) Create Physical Map of Region (Genomic “walk”) oep Use 15AH and 20K markers to gain “entry” into genomic region -Make probes using RAPD PCR bands from each -Probe genomic library to isolate clones 134 and 32 -Use ends of these to isolate contiguous clones (“walk”) -Stop when two directions of walk meet If genome sequence is available, don’t have to walk since you know sequence of interval between markers 2) Create Physical Map of Region (Genomic “walk”) oep Clone 134F10 failed the “deletion test” -see if all of clone is missing in deletion of oep genomic region -if not missing, then some of clone’s DNA is from other region -suggests clone is chimeric (contains different parts of genome) -would be disaster to continue “walking” from chimeric clone could jump to entire new (irrelevant) region or new chromosome 3) Recombinant Fine Mapping oep -Subclone 14 and 240 into cosmids -Use ends to make STS’s (need spaced sequence info) -Use sequence to identify additional polymorphic markers SSCPs and CAPS -Go back to previously identified recombinants (1 on left, 5 on right) -Use new markers to map recombination events e.g. 46T7 has allele of “strain 2”, so recombination is b/w 46T7 and oep -Therefore oep is between 46T7 and 32T7 4) Identify the Gene oep -Use genomic DNA to probe cDNA library from stage of interest (223 cDNAs!) -Find out that these represent 13 “classes” of cDNA -Use in situ hybridization to see if any expression patterns fit predictions -Sequence mutant genomic DNA to identify potential bp changes that are responsible for mutant phenotype -Rescue with in vitro synthesized RNA -Gold Standard for Gene ID: sequence point mutations and rescue mutant defect with transcript How would things be changed today? -High density polymorphism map produced so don’t need to search for polymorphic markers -Genome sequence being completed so don’t need to walk -Large scale EST (cDNA) sequencing so know transcript distribution and candidate genes (at least those that are correctly annotated!) -Can use morpholinos (RNAi in other species) to test candidate transcripts -Whole genome sequencing becoming helpful for identifying mutations http://www.hapmap.org/ Goal: Determine the existing human haplotypes with a defined set of SNPs Long Term Goal: Associate haplotypes with phenotypes for -cloning disease genes -understanding genetics of complex traits -pharmacogenomics