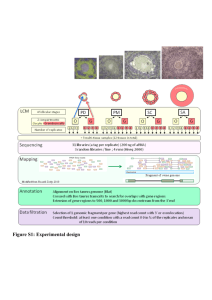
Intro: sequencing and the data deluge
advertisement

MCB3895-004 Lecture #21 Nov 20/14 Prokaryote RNAseq Today: • Building off last lecture, we will use reference alignment methods to understand differential gene expression in prokaryotes • Use Bowtie2 for alignment • Use Edge-pro for determining transcript abundance Experiment: • Compare E.coli K-12 grow in glucose minimal medium aerobically vs. anaerobically • Aerobic datasets: SRR922260 • Anaerobic datasets: SRR922265 • All sequenced using Illumina GAIIx, 2x36bp PE Basic idea of RNAseq • One way to analyze a transcriptome (i.e., all the mRNA molecules) is to count the number of transcripts from each gene • More transcripts implies more activity of that gene • Improvement over previous technology (microarrays) that required some knowledge of what genes to look for and were less sensitive Problems: 1. How to compare short genes to long ones? • Short genes will have fewer reads mapping to them by random chance 2. How to compare genes from different genomes with different sampling intensity? • Transcripts sampled more deeply will have more reads mapping to them RPKM • "Reads per kilobase per million" • RPKM normalizes for both gene length and sampling intensity • RPKM = [# of mapped reads]/[length of transcript in kb]/[million mapped reads] • Allows genes to be compared to each other • Allows transcription to be compared between transcriptomes RNAseq software • Many packages exist for comparing transcriptomes • Most are tailored towards eukaryotes • Emphasis on finding splice variants (not in bacteria) • Do not account for overlapping genes (common in bacteria, rare in eukaryotes) Generalized scheme for RNAseq 1. Map reads to reference genome 2. Count reads mapping to each gene 3. Normalize for gene length and sampling depth (i.e., calculate RPKM) 4. Statistically compare test and control sample sets (a topic in itself, not covered in depth here) EDGE-pro • The software we will use is EDGE-pro • Installed on server in /opt/bioinformatics/EDGE_pro_v1.3.1/ • Tailored for prokaryotes • Magoc et al. (2013) Evolutionary Bioinformatics 9:127-136 • http://ccb.jhu.edu/software/EDGE-pro/ EDGE-pro outline 1. 2. 3. 4. 5. Use Bowtie2 to map reads Calculate per base coverage Assign per gene coverage Disambiguate overlapping genes Calculate RPKM for each gene Running EDGE-pro • syntax: $ perl /opt/bioinformatics/EDGE_pro_v1.3.1/edge.pl -g [.fna name] -p [.ppt name] -r [.rnt name] -u [.fastq 1 name ] -v [.fastq 2 name] -s /opt/bioinformatics/EDGE_pro_v1.3.1/ • • • • • • -g: reference .fna file name -p: reference .ptt file name -r: reference .rnt file name -u: .fastq file name to map -v: .fastq file pairing with that specified by -u, if exists -s: location where program lives • e.g.: $ perl /opt/bioinformatics/EDGE_pro_v1.3.1/edge.pl -g NC_000913.fna -p NC_000913.ptt -r NC_000913.rnt -u SRR922260_1.fastq -v SRR922260_2.fastq -s /opt/bioinformatics/EDGE_pro.v1.3.1/ EDGE-pro: results • One nice thing about EDGE-pro is that it runs many scripts all by itself • A "wrapper" or "pipeline" is something that bundles different programs altogether • Many of the output files are from bowtie2, some are from EDGE-pro itself • Note: make sure that you have enough space in your account for these files • The RPKM data are located in "out.rpkm_0", which is a tab-delimited table listing the reads mapped to each predicted transcript Comparing conditions • There are many different ways to compare test and control conditions • This is outside of the scope of this class • The RPKM values generated by EDGE-pro can be reformatted to be input • EDGE-pro contains a script that will do this for DESeq, one of the most popular • Generally multiple replicates should be considered for each condition EDGE-pro comparison • The EDGE-pro paper suggests an easy heuristic for transcriptome comparison: 1. Average RPMK values from treatment replicates 2. Determine the RPMK fold change between test and control treatments using simple division 3. Only keep results >4-fold different A reference genome quirk: • EDGE-pro requires the standard .fna genome file and .ptt and .rnt files that list gene locations on the chromosome • Unfortunately only available from the old version of the NCBI ftp server • Location for today: ftp://ftp.ncbi.nlm.nih.gov/genomes/ Bacteria/Escherichia_coli_K_12_subs tr__MG1655_uid57779/ Today's assignment • Use EDGE-pro to calculate RPMK values for the E.coli K-12 RNAseq transcriptomes generated under aerobic (SRR922260) and anaerobic (SRR922265) conditions • Write a short perl script to calculate the recommended EDGE-pro comparison • Only one replicate so no averaging needed • Report 4-fold overrepresented genes in aerobic treatment • Report 4-fold overrepresented genes in anaerobic treatment