A new way of ordering endophenotypes for relevance to a disease
advertisement

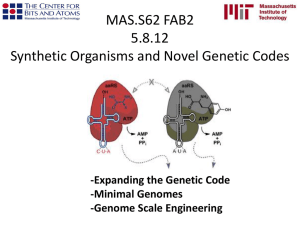
The search process: A way to rank endophenotypes (e) for relevance to a disease (i) phenotype “What is an endophenotype?” you ask. Great Question. Some Candidates. THESE WERE PLUCKED OUT OF 11,000 TRAITS: Behavioral/Neurocognitive Beck Depression Inventory II EPQ Neuroticism Declarative Memory (CVLT Recognition) Working Memory (Digit Span Forward) Working Memory (Letter-Number Span) Penn Facial Memory (Immediate) Penn Facial Memory (Delayed)Attention (CPT hits) Attention (Trails A) Penn Emotion Recognition Neuroimaging Ventral diencephalon volume Parietal hyperintensity volume Hippocampus volume Pallidum volume Cerebellar white matter volume Frontal hyperintensity volume Corticospinal tract (FA) Subcortical hyperintensity volume Superior parietal gyrus thickness Thalamus proper volume Transcriptional RNF123 (3p24) PDXK (21q22) ZFP64 (20q13) RWDD2A (6q14) B4GALT7 (5q35) MARK2 (11q12) GADD45A (1p31) PIGN (18q21) HTT (4p16) ABHD12 (20p11) Glahn et al., 2012 High Dimensional Endophenotype Ranking in the Search for Major Depression Risk Genes Endophenotypes are: • • • • NOT A CLINICAL SUBTYPE HERITABLE Trait-related deficit An INTERMEDIATE phenotype that stresses the genetic link • State-INDEPENDENT (present whether or not disease is active) • Found WITH GREATER FREQUENCY (as compared to general population) in related members of affected individuals and affected individuals. • The idea is that these will be less heterogeneous than clinical measures Braff et al., 2007 Deconstructing Schizophrenia Gottesman, 2003 Psychiatric Endophenotypes and the development of valid animal models Endophenotype Ranking Value (ERV): An Index of genetic utility (standardized genetic covariance) of e for i ERVie 2 i h h g 2 e i = illness e= endophenotype ρg= genetic correlation of i & e About the Endo Ranking Value • ERV’s will vary between 0 and 1 – higher values ≈ stronger shared genetic influence • ERV balances the strength of the genetic signal for the e and the strength of its relation to the i • Advantage: large #s of potential es can be assessed before conducting molecular genetic analyses- useful in guiding the search. ERV ingredients ERVie h 2 p g g2 2 p additive g g(i, e) e 2 e g (i) g (e) h h g 2 e Broad genetic heritability is the genotypic variance divided by the phenotypic variance h h 1 h 1 h 2 2 e i 2 i 2 i Phenotypic correlation derived from the genetic and environmental correlations (obtained by using all pedigree info avail with a multivariate normal threshold model for combined dichotomous/continuous traits Let’s take a look at what an ERV looks like when you change the heritability values of e and i… The Example Study: recurrent Major Depressive Disorder (rMDD) • N= 1122 from 71 extended pedigrees • 11,000 traits • FIRST: they rank their endophenotypes • SECOND: they check the utility of these rankings with bivariate linkage analyses (rMDD + endophenotypes) Bivariate Linkage Analysis (to improve power to detect) • Exploits the genetic covariance between the endophenotype and the illness to improve the power to localize QTLs (quantitative trait locus) and to detect QTL-specific pleiotropic effects • Compute maximum likelihood estimates of allele frequencies • Matrices of empirical estimates of identity-by-descent allele sharing at points throughout the genome for every relative pair were computed • Once genome-wide significant localization was made, formal single degree of freedom likelihood ratio tests for pleiotropy were performed to test the specific hypothesis that a QTL at that location influenced a given endophenotype risk. QTLs (quantitative trait loci) found • 4p15 (using univariate linkage) – Bilateral ventral p= 4.7 x 10-5 – RNF123 p= 0.0010 – Diencephalon volume p= 0.0290 – BDI-II p= 0.1170 (trend) – The linkage analysis provides further validation that the endophenotype identification strategy can be used to find genetic influence – Now these can be molecularly analysed. Testing for Pleiotropic Effects THESE WERE PLUCKED OUT OF 11,000 TRAITS: Top Results Behavioral/Neurocognitive Beck Depression Inventory II EPQ Neuroticism Declarative Memory (CVLT Recognition) Working Memory (Digit Span Forward) Working Memory (Letter-Number Span) Penn Facial Memory (Immediate) Penn Facial Memory (Delayed)Attention (CPT hits) Attention (Trails A) Penn Emotion Recognition Neuroimaging Ventral diencephalon volume Parietal hyperintensity volume Hippocampus volume Pallidum volume Cerebellar white matter volume Frontal hyperintensity volume Corticospinal tract (FA) Subcortical hyperintensity volume Superior parietal gyrus thickness Thalamus proper volume Transcriptional RNF123 (3p24) PDXK (21q22) ZFP64 (20q13) RWDD2A (6q14) B4GALT7 (5q35) MARK2 (11q12) GADD45A (1p31) PIGN (18q21) HTT (4p16) ABHD12 (20p11) Glahn et al., 2012 High Dimensional Endophenotype Ranking in the Search for Major Depression Risk Genes Varying heritability affecting the ERV value. ht Mark + Tim