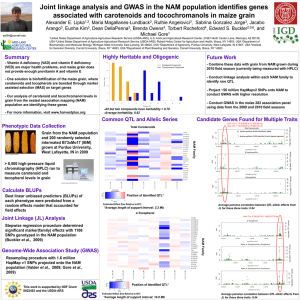
7 GEM Cooperators Me..
advertisement
Genetic Architecture of Kernel Composition in the Nested Association Mapping (NAM) Population Sherry Flint-Garcia USDA-ARS Columbia, MO Outline Development of NAM Population Kernel Composition Joint Linkage Mapping Genome-Wide Association Mapping Linkage-Based QTL Mapping “Genome Scan” Identify genomic regions that contribute to variation and estimate QTL effects 140 130 120 110 100 90 80 70 60 50 40 30 20 Parent 2 0 Parent 1 10 Position (cM) 9 6 5 4 3 2 1 b1520 b2077 b1225 b2248 m231 b2277 0 u1552 F2 population Composite Interval Mapping 7 u1622 F1 8 LOD Score Genotype Phenotype Linkage (QTL) Mapping Genome scan Structured population High power Low resolution Analysis of 2 alleles Association Mapping Candidate gene testing Unstructured population Low power High resolution Analysis of many alleles Nested Association Mapping Structured families nested within an unstructured population High Power High Resolution Analysis of many alleles CM37 R4 K148 Ky228 NC344 DE-3 Oh7B K4 NC360 A682 NAM Founders NC362 CI91B NC262 NC342 NC364 NC258 CMV3 Mt42 Mo17 CI187-2 Yu796-NS Mo45 B97 A556 CI3A Hi27 Mo47 CO106 W401 NC222 Mo46 B77 B103 W117HT Tzi16 Tzi25 DE811 B105 MS153 DE1 SD40 NC290A A641 A214N B164 NC236 DE-2 CM7 B57 N7A N28HT NC250 H100 STIFF STALK H84 I205 C123 CO109 ND246 B64 H105W A635 A632 C103 CO125 B79 H91 A634 B68 B84 B14A Hy Ky21 A661 CI21E B76 CM174 WD CM105 A554 B104 B75 38-11 B37 Os420 NC260 MS71 Mo1W NC328 Mo44 A679 NC294 NC368 B109 N192 CH9 NC292 WF9 SD44 OH43 A239 A188 Pa762 VA26 Ky226 C49A Va102 Oh43E Va35 Va14 Oh40B NC372 NC310 B10 B46 Pa880 T8 NC324 NC330 NC322 NC308 NC312 NC306 NC268 H49 B73 NC326 NC314 W64A Pa875 A619 R229 A680 B73Htrhm R168 Va59 C49 A654 W153R A659 Va17 CI-7 R177 Va22 W22 H95 W182B Va99 PA91 H99 M14 CI90C NON STIFF STALK 33-16 Va85 CH701-30 NC33 VaW6 4226 NC232 L317 B115 R109B MoG I137TN K55 CI66 CI44 NC230 81-1 CI31A MEF 156-55-2 CI64 IL677A M162W K64 E2558W Ia5125 N6 IA2132 T234 CML52 CML69 CML103 L578 CML14 IL14H P39 SC357 IL101 CML38 SWEET B52 CML287 EP1 Tzi11 NC366 CML108 F2 F7 SC213R CO255 CML9 GT112 CML61 NC238 CML254 CML5 T232 CML314 GA209 CML264 CML258 CML10 B2 CML11 CML45 CML341 CML261 CML331 CML332 Sg1533 SG18 Mo24W IDS28 CML238 CML321 A6 Ki14 F2834T M37W CML157Q SA24 F44 Ki11 Ki2021 F6 Ki44 4722 I-29 Oh603 Ki43 POPCORN CML328 Ki21 D940Y HP301 CML277 CML247 CML322 U267Y MS1334 IDS69 IDS91 TROPICALSUBTROPICAL Mp339 CI28A Q6199 NC340 Ki2007 CML323 CML92 CML228 Ki3 NC338 TZI18 NC300 NC358 A272 NC356 NC302 NC354 CML218 CML77 NC332 Tx303 NC318 SC55 A441-5 NC334 CML154Q NC370 TZI10 NC264 Ab28A CML220 Tzi9 NC350 NC320 TX601 CML349 CML158Q NC304 CML91 CML333 Tzi8 CML311 Mo18W MIXED 0.1 Based on 89 SSR loci CML281 NC296A NC346 parvi-03 NC336 NC296 NC352 NC348 NC298 parvi-30 ssp. parviglumis parvi-14 parvi-49 parvi-36 Flint-Garcia, et al. (2005) Plant J. NAM Development B97 Linkage Current genetic map consists of: CML103 CML228 CML247 CML277 CML322 4699 RILs 1106 SNP loci CML333 CML52 CML69 Association Hp301 Ki11 Ki3 Il14H Ky21 Average marker density - one marker every 1.3 cM M162W M37W Mo18W MS71 NC350 NC358 Oh43 Oh7B P39 Tzi8 Tx303 25 DL SSD Yu, et al. (2008) Genetics; McMullen, et al. (2009) Science Kernel Composition in NAM Fiber Starch Amylose Zeins Amylopectin Protein Oil Amino Acid Profiles Fatty Acid Profiles The Phenotypic Data 7 locations of NAM – 2006: MO, NY, NC, PR, FL 2007: MO, NY Self pollinated seed samples NIR analysis for starch, protein, and oil content (% kernel - dry matter basis) Two sweet corn families excluded >6000 rows per location Phenotypic Data Statistics Heritability (23 Families) Trait Correlations r H2 Starch 0.85 Protein 0.83 Oil 0.86 Starch Protein Protein Oil -0.65 -0.40 0.32 NAM Analysis in SAS Permutations for selection thresholds ~10-5 Joint stepwise regression; Proc GLMSelect Family main effect & markers within families Final model; Proc GLM Estimate effects (P = 0.05) Genome Scan; Proc Mixed Maximum likelihood with background cofactors Epistasis; all (611,065) pair-wise combinations NAM Kernel Quality Architecture 80 70 60 LOD 50 Starch Protein Oil 40 30 20 10 0 Trait N R2(family) R2(QTL) Starch Protein Oil 21 26 22 28.7 25.8 44.5 58.1 59.9 69.0 R2(QTL+family) 59.1 61.0 69.7 No Epistasis Observed at the NAM Level Sig. Alleles N (P = 0.05) Starch Protein Oil 180 206 174 Min Max (%) (%) % -0.5 -0.3 -0.1 0.0 0.1 0.3 0.5 -0.62 0.65 -0.38 0.34 -0.12 0.21 62.3 63.3 63.4 63.8 63.9 64.8 65.2 65.3 65.5 66.2 66.3 66.8 67.3 67.3 67.5 67.5 67.6 67.6 67.6 67.8 68.2 68.7 68.9 HP301 CML247 Ki11 CML322 Ki3 CML333 MS71 CML228 Tzi8 CML69 Oh7B NC350 Ky21 CML52 CML277 Tx303 M37W Mo18W M162W NC358 B97 Oh43 CML103 1_20.1 1_70.3 1_146 2_210.9 2_277.2 2_306.1 3_420.5 3_478.8 3_503.4 4_650.2 5_724.7 5_753.5 6_819.4 6_836.8 7_993.1 7_1013 8_1102.7 8_1141 9_1224.4 9_1245.5 10_1338.2 Starch Additive Allelic Effects -0.52 -0.32 -0.30 -0.34 -0.31 -0.55 -0.45 0.56 -0.35 -0.33 0.51 -0.25 -0.30 B73 % -0.3 -0.2 -0.1 0.0 0.1 0.2 0.3 12.3 12.5 12.6 12.7 12.7 12.9 13.1 13.2 13.3 13.3 13.6 13.8 13.8 14.0 14.0 14.2 14.2 14.4 14.6 14.7 14.8 15.2 15.3 B97 NC358 CML277 Mo18W M162W Tx303 Ki3 Oh43 HP301 CML103 CML69 CML333 Ky21 MS71 CML228 Ki11 CML52 M37W CML247 CML322 Oh7B NC350 Tzi8 -0.10 -0.10 -0.18 0.10 -0.12 0.15 -0.27 -0.26 -0.46 0.25 0.25 -0.24 -0.40 -0.41 0.29 -0.62 -0.42 -0.28 -0.25 -0.37 -0.20 0.20 -0.44 -0.33 0.19 -0.33 -0.23 -0.25 -0.22 -0.26 0.28 0.23 -0.40 -0.47 -0.21 0.24 0.27 0.26 -0.26 -0.20 0.16 -0.25 0.11 0.15 -0.16 0.15 0.15 0.10 -0.12 -0.13 -0.34 0.32 0.18 0.21 -0.20 -0.19 0.33 0.16 -0.38 0.17 0.15 -0.15 0.12 0.34 0.13 -0.15 0.13 0.14 -0.19 -0.11 0.12 -0.15 -0.13 -0.15 -0.20 0.20 -0.10 0.15 -0.19 -0.36 0.12 -0.15 0.10 0.17 0.16 -0.13 -0.18 0.15 0.13 0.10 0.17 -0.11 0.13 0.11 -0.11 0.23 0.20 -0.13 0.14 -0.17 0.20 -0.16 0.14 -0.13 0.17 0.25 0.18 0.23 0.32 -0.12 0.20 -0.22 -0.11 -0.17 0.19 0.13 0.15 -0.13 0.22 -0.26 0.13 -0.12 -0.13 0.19 -0.17 0.18 0.13 -0.14 -0.20 -0.17 0.18 0.22 0.24 0.15 0.11 0.16 0.21 -0.15 -0.23 -0.24 -0.12 0.16 -0.12 -0.18 0.19 -0.45 -0.34 -0.27 0.23 0.22 0.24 -0.38 0.20 % -0.15 -0.10 -0.05 0.00 0.05 0.10 0.15 0.18 -0.21 0.19 -0.25 -0.46 0.40 -0.20 -0.47 0.27 -0.21 -0.32 0.31 0.28 -0.52 -0.25 0.17 -0.27 -0.30 0.34 -0.22 -0.25 -0.25 0.22 0.40 -0.47 0.23 -0.27 -0.27 0.31 0.25 -0.23 -0.29 0.33 0.30 -0.34 -0.28 0.28 0.20 0.21 0.25 0.20 0.25 -0.29 0.22 -0.29 -0.26 -0.24 -0.35 -0.23 -0.23 0.36 0.23 -0.43 -0.55 0.34 -0.31 0.38 0.26 0.19 -0.55 -0.21 0.35 0.22 -0.27 0.30 -0.34 -0.37 0.43 0.27 -0.23 -0.20 -0.25 -0.32 0.27 -0.22 0.43 3.5 3.6 3.6 3.6 3.9 3.9 4.0 4.0 4.1 4.2 4.2 4.3 4.4 4.4 4.6 4.6 4.7 5.1 5.2 5.3 5.3 5.4 5.5 0.36 ^ 1_33.1 1_116.5 1_141.9 1_199.4 2_266.6 2_282.2 2_308.5 3_414.1 3_462.1 4_571 4_592 4_642.9 5_668 5_692.6 5_715.3 5_741.9 6_841.5 6_869.5 7_969 8_1135.1 9_1240.3 10_1355.8 10_1399.3 -0.27 0.29 -0.18 -0.11 -0.16 10_1353.5 10_1324.2 0.18 0.12 -0.19 -0.18 -0.14 -0.11 0.12 -0.14 -0.27 0.11 -0.23 -0.31 -0.23 0.20 -0.14 -0.11 0.24 0.14 -0.13 -0.11 -0.12 0.15 -0.16 0.14 -0.12 0.11 -0.22 0.13 -0.18 -0.13 0.13 -0.11 0.11 -0.25 0.20 -0.19 0.12 0.10 0.19 -0.22 -0.12 0.10 0.12 -0.11 0.15 0.23 -0.16 -0.22 0.15 -0.20 B73 0.10 0.21 0.15 -0.12 -0.13 0.15 -0.13 0.10 -0.13 -0.22 -0.18 0.13 -0.18 -0.22 0.17 0.12 0.15 0.39 0.38 0.19 -0.19 0.29 0.26 Oil -0.18 -0.15 -0.17 -0.15 -0.11 -0.12 -0.34 9_1256.6 9_1222.6 8_1149.3 8_1128.9 -0.13 0.11 -0.26 -0.19 -0.25 0.50 0.20 -0.32 -0.50 -0.22 -0.28 -0.39 0.39 0.36 0.29 -0.25 -0.24 -0.26 -0.23 0.43 -0.21 -0.25 -0.28 0.42 -0.61 -0.33 0.28 0.65 -0.23 -0.41 -0.21 -0.32 -0.22 -0.20 ^ 0.15 -0.20 -0.11 -0.18 0.16 -0.19 8_1106.7 -0.15 -0.17 -0.18 0.15 7_1013 7_987.6 6_914.7 0.18 0.22 -0.10 6_862.1 6_836.8 6_819.2 5_756.6 5_723.4 4_595.3 4_575.4 3_503.4 3_420.5 2_355.8 2_297.3 2_255.7 1_162.2 1_137.6 1_56.5 -0.27 -0.22 -0.34 -0.38 0.50 B73 Protein -0.38 -0.26 -0.20 -0.26 -0.28 0.47 Oh43 M37W Ky21 CML247 NC358 Tx303 CML277 B97 Mo18W MS71 HP301 CML103 M162W CML52 Ki3 CML69 Ki11 CML228 Tzi8 Oh7B NC350 CML322 CML333 0.11 0.05 -0.10 -0.06 -0.09 0.06 -0.05 0.04 -0.07 -0.08 -0.04 0.05 0.06 0.06 -0.11 -0.05 0.10 0.18 -0.06 -0.11 -0.06 0.07 -0.05 0.10 0.05 0.05 -0.05 0.05 -0.11 0.08 0.11 -0.09 -0.08 0.06 0.06 -0.07 0.06 -0.12 0.09 0.11 -0.07 0.07 0.07 0.21 0.07 0.05 -0.05 -0.09 -0.07 0.05 -0.07 0.06 -0.06 0.04 0.05 -0.06 -0.05 0.05 0.06 -0.10 -0.06 0.05 0.07 -0.10 0.05 0.06 -0.11 0.10 0.07 -0.05 0.08 0.06 0.06 -0.10 -0.05 0.07 -0.07 -0.05 -0.05 0.05 -0.07 0.07 0.07 0.10 -0.11 0.12 -0.06 0.08 0.04 0.08 0.07 0.09 -0.07 0.08 -0.05 -0.08 -0.04 -0.06 0.07 -0.09 0.07 0.12 0.04 -0.07 0.06 0.07 0.06 0.05 0.10 0.10 0.13 -0.07 0.05 0.12 0.09 0.07 0.12 0.12 0.08 0.06 -0.06 -0.05 0.12 0.12 -0.10 -0.06 0.12 -0.10 0.06 0.06 -0.05 0.05 0.11 0.11 0.07 0.08 -0.06 0.06 0.10 0.06 0.08 -0.09 -0.06 0.12 0.05 0.11 0.05 0.06 0.12 -0.05 -0.06 0.06 0.06 -0.11 0.08 -0.10 -0.10 0.16 0.06 0.14 -0.10 0.10 -0.04 0.10 0.09 0.10 -0.06 -0.06 0.05 0.10 0.08 0.05 0.11 -0.04 Validation Efforts Near Isogenic Lines (NILs) Genome Scan Association Analysis Candidate Genes Association Analysis Candidate Floury1 Opaque2 Modifier/Mucronate Brittle Endosperm1 DGAT1-2 Waxy1 Marker m221 m261 m619 m708 m968 Chr. 2 2 5 6 9 Dist. 266.6 297.3 724.7 841.5 1224.4 Fine Mapping Trait Oil Oil Oil Oil Protein Starch Allele Tx303 CML322 CML228 Tx303 CML103 Tzi8 Marker m707 m401 m94 m565 m657 m353 Chr. 6 3 1 5 5 3 Dist. (cM) 841.5 462.1 116.5 692.6 756.6 420.5 Jason Cook Trait Oil Protein Starch Oil Starch Estimated Effect (%) 0.21 0.11 0.11 - 0.12 0.21 0.43 Genetic vs. Physical Distance 80 70 Joint Linkage Mapping - Oil 60 40 30 20 10 0 0 100 200 300 400 500 600 700 800 900 1000 1100 1200 1300 1400 Genetic Distance (cM) 90 80 Joint Linkage Mapping - Oil 70 60 LOD LOD 50 50 40 30 20 10 0 - 500,000,000 1,000,000,000 Physical Distance (bp) 1,500,000,000 2,000,000,000 Genome Wide Association (GWAS) 90 80 Joint Linkage Mapping - Oil 70 LOD 60 50 40 30 20 10 0 100.0 90.0 500,000,000 1,000,000,000 1,500,000,000 2,000,000,000 80 70 GWAS - Oil 80.0 60 BPP BPP 70.0 50 60.0 50.0 40 40.0 30 30.0 20 20.0 10 10.0 0.0 0 - 500,000,000 1,000,000,000 1,500,000,000 Physical Distance (bp) 1.6 Million HapMap v1 SNPs projected onto NAM Bootstrap (80%) sampling to test robustness 2,000,000,000 Chr. 6 Oil Candidate: DGAT1-2 Oil Effect (%) 0.25 0.20 0.15 0.10 0.05 0.00 -0.05 -0.10 M37W Oh43 CML103 CML322 HP301 MS71 B97 NC358 CML69 Ki11 CML277 CML247 Mo18W CML333 CML52 NC350 Ki3 CML228 Tzi8 M162W Oh7B Ky21 Tx303 Encodes acyl-CoA:diacylglycerol acyltransferase Fine mapped by Pioneer-Dupont Zheng, et al. (2008) Nature Genetics High parent = 19% oil High allele = 0.29% additive effect DGAT is the largest effect kernel quality QTL in NAM 4.4% 5.3% 3.6% 3.9% Phenylalanine insertion in the C-terminus of the protein DGAT 1-2 (Chr6: 105,013,351-105,020,258) M1 M2: Phe Insertion M3 Marker Trait M1 Oil M2 Oil M3 Oil M4 Oil M4 Starch M5 Oil M5 Starch Population 282 Assn. 282 Assn. NAM 282 Assn. NAM NAM NAM M4 Analysis Method MLM (Q+K) MLM (Q+K) GWAS - Bootstrap MLM (Q+K) GWAS - Bootstrap GWAS - Bootstrap GWAS - Bootstrap BPP 31 51 67 11 M5 P-Value 1.2E-04 9.9E-04 4.3E-05 - Effect 0.18 0.16 0.18 0.19 -0.38 0.13 -0.31 NAM Population: 24 Total HapMap.v1 SNPs in DGAT Association Panel: 2 Total 55K SNPs in DGAT DGAT 1-2 (Chr6: 105,013,351-105,020,258) M1 M2: Phe Insertion Oil Effect (%) 0.25 0.20 0.15 0.10 0.05 0.00 -0.05 -0.10 M37W Oh43 CML103 CML322 HP301 MS71 B97 NC358 CML69 Ki11 CML277 CML247 Mo18W CML333 CML52 NC350 Ki3 CML228 Tzi8 M162W Oh7B Ky21 Tx303 M3 M4 M5 = B73 Allele = Non-B73 Allele ? What’s Next for NAM? NextGen sequencing of the 5000 NAM RILs Potentially 30-50 Million SNPs Identify very precisely where recombination events are in the mapping population. This will VASTLY improve the mapping resolution of NAM and GWAS. Conclusions Genetic Architecture of Kernel Quality Traits Governed by many QTL (N = 21-26) Many QTL in common with prior studies Effect sizes are small to moderate Allele series are common Genome Wide Association Studies (GWAS) Results confirm many QTL and candidate genes Resolution will improve with more markers on NAM RILs (define recombination events) What Does This Mean To You? Identifying Functional Markers for MAS (Distantly) Linked markers not accurate Parent Selection = Allele Mining Valuable alleles are often masked. Selection for specific alleles is more accurate than selecting based on parental phenotype. Acknowledgements NSF Maize Diversity Project www.panzea.org Syngenta Joe Byrum & Kirk Noel GEM Allelic Diversity Project 250 Races B47 (SS) PHZ51 (NSS) Allele Library 2500 lines Genome Wide Association Analysis “mini-NAM” Allele Mining