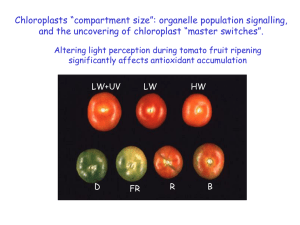
PowerPoint 1
advertisement

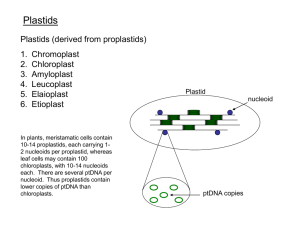
PCB6528 Plant Cell and Developmental Biology Spring 2012 Organelle genomes, gene expression and signaling Christine Chase – 2215 Fifield Hall – 352-273-4862 cdchase@ufl.edu Organelle genomes Organelle gene expression processes Organelle-to-nucleus signaling (retrograde regulation) Objectives - Organelle genomes: Describe the organization and coding content of plant plastid and mitochondrial genomes Discuss the similarities and differences between the plastid and plant mitochondrial genomes with respect to organization and evolution Explain why plastid or mitochondrial genome coding content is not necessarily identical between plant species Discuss the possible reasons that plant organelles retain genomes at all Describe the process of plastid genome transformation Discuss the utility and applications of plastid transformation and provide some specific examples Organelle genomes Small but essential genomes Multiple organelles per cell; multiple genomes per organelle (20 – 20,000 genomes per cell, depending on cell type) Organized in nucleo-protein complexes called nucleoids Non-Mendelian inheritance; usually but not always maternally inherited in plants Encode necessary but insufficient information to elaborate a fully functional organelle Many nuclear gene products required for organelle function translated on cytosolic ribosomes & imported into the organelles plant mitochondria also import tRNAs needed for a complete set! Considerable cross-talk between nuclear and organelle genetic systems Comparative sizes of plant genomes Genome Size in bp Arabidopsis thaliana 1.4 x 10 8 3.7 x 10 5 nuclear Arabidopsis thaliana mitochondria Arabidopsis thaliana plastid Zea mays nuclear Zea mays mitochondria Zea mays plastid 1.5 x 10 5 2.4 x 10 9 5.7 x 10 5 1.4 x 10 5 Organelle genomics & proteomics Target P prediction analysis of the complete Arabidopsis nuclear genome sequence (Emanuelsson et al., J Mol Biol 300:1005)says ..... ~ 10% of the Arabidopsis nuclear genome (~2,500 genes) encode proteins targeted to the mitochondria ~ 14% of the Arabidopsis nuclear genome (~3,500 genes) encodes proteins targeted to the plastid So 25% of the Arabidopsis nuclear genome is dedicated to organelle function! Proteome reflects metabolic diversity of these organelles, both anabolic and catabolic Endosymbiont origin of organelles * ** Original basis in cytology Confirmation by molecular biology α proteobacteria as closest living relatives to mitochondria Cyanobacteria closest living relatives to plastids Archaebacteria considered to be related to primitive donor of the nuclear genome * * * [Gillham 1994 Organelle Genes and Genomes] Chimeric origin of eukaryotic nuclear genomes Genes per category among 383 eubacterial- & 111 archeaebacterialrelated genes in the yeast nuclear genome Esser et al. 2004 Mol Biol & Evol 21:1643 Evolution of the eukaryotic genomes Reduced coding content of organelle genomes •Functional gene transfer to nucleus with protein targeted back to organelle •Functional re-shuffling - organelles replace prokaryotic features with eukaryotic, “hybrid” or novel features Evolution of mitochondrial genome coding content Genome Protein coding genes Rikettsia prowazekii 832 (smallest proteobacterial genome) Reclinomonas americana 62 Marchantia polymorpha 64 Arabidopsis thaliana mitochondria 57 mitochondria (protozoan; most mitochondrial genes) mitochondria 1.9 x 10 5 bp (liverwort, non-vascular plant ) 3.7 x 10 5 bp (vascular plant) Homo sapiens mitochondria 13 Evolution of plastid genome coding content Genome Protein coding genes Synechococcus (cyanobacteria) 3,300 Paulinella chromatophora 867 photosynthetic body (endosymbiont cyanobacteria) Porphyra purpurea plastid (red alga) Chlamydomonas reinhardtii plastid (green alga) Marchantia polymorpha plastid (liverwort, non-vascular plant) Arabidopsis thaliana plastid (vascular plant) Epifagus virginiana plastid (non-photosynthetic parasitic plant) 209 63 67 71 42 Functional gene transfer from organelle to nuclear genome • Gene by gene • Likely occurs via RNA intermediates • Evidence for frequent and recent transfers in plant lineage • Results in coding content differences among plant organelle genomes • What is required for a functional gene relocation from organelle to nucleus? • How would we know this occurs via RNA intermediates? Functional gene transfer: Recent repeated transfers of the plant mitochondrial rps10 to the nucleus •Southern blot hybridization of total cellular DNA samples • Mitochondrial nad1 and rps10 probes • Shading = taxa with no hybridization to rps10 • Bullets = taxa with confirmed nuclear copy of rps10 [Adams et al. Nature 408:354] • Why is there no hybridization of rps10 probes to DNA samples with confirmed nuclear copy of rps10? (Hint: How are the relative genome copy numbers and sizes exploited in this screen?) •What is the purpose of the nad1 probe? •What are the consequences of these events with respect to plant mitochondrial genome coding content? Reduced plastid genome content in nonphotosynthetic plastids: Parasitic plants with 70-20 kb plastomes have lost photosynthetic genes, some ribosomal proteins & some tRNAs Essential tRNA hypothesis: [Barbrook et al. Trends in Plant Sci. 11:101] Plastid tRNAs needed to support mitochondrial function • tRNA-Glu as precursor for the synthesis of heme for mitochondrial respiratory electron transfer • tRNA-Met imported into mitochondria for mitochondrial protein synthesis Epifagus virginiana (beechdrops) A non-phoptosynthetic, parasitic plant Has a plastid genome of 71 kb encoding 7 tRNAs and 2 ribosomal proteins http://2bnthewild.com/plan ts/H376.htm Land Plant Plastid Genome Organization Physical map (e. g. restriction map or DNA sequence) indicates a 120-160 kb circular genome Large inverted repeat (LIR) commonly 20-30 kb •Large single copy (LSC) region •Small single copy (SSC) region Active recombination within the LIR Expansion and contraction of LIR • Primary length polymorphism among land plant species • 10-76 kb Some conifers and legumes have very reduced or no LIR Inversion polymorphisms within single copy regions mediated by small dispersed repeats Plastid genome organization (Maier et al. J Mol Biol 251:614) Plastid genes in operons (from Palmer [1991] in Cell Culture and Somatic Cell Genetics of Plants, V 7A. L Bogorad and IK Vasil eds. Academic Press, NY, pp 5-142) Recombination across inverted repeats leads to inversions rps15 ndhF trn N trn N ndhB ndhB rps19 rps19 rpl22 psbA How can these inversion isomers be detected? ndhF trn N ndhB rps19 psbA rps15 trn N ndhB rps19 rpl22 Fiber FISH of tobacco plastid DNA [Lilly et al. Plant Cell. 13:245] Structural Plasticity of cpDNA Molecules from Tobacco, Arabidopsis, and Pea [Lilly et al. Plant Cell. 13:245] Structural Plasticity of cpDNA Molecules from Tobacco, Arabidopsis, and Pea Table 1. Frequency of Different cpDNA Structures across All Experiments in Three Species No. of Observations Structurea Arabidopsis Tobacco Pea Circular 126 (42%) 524 (45%) 59 (25%) Linear 68 (23%) 250 (22%) 85 (36%) Bubble/D-loop 25 (8%) 67 (6%) 5 (2%) Lassolike 34 (11%) 115 (10%) 21 (9%) Unclassifiedb 44 (16%) 203 (17%) 66 (28%) a Each classification represents all molecules of that type regardless of size. b DNA fibers that were coiled or folded and could not be classified [Lilly et al. Plant Cell. 13:245] Plastid genome coding content Chloroplast Genome Database: http://chloroplast.cbio.psu.edu/ (Cui et al., Nucl Acids Res 34: D692-696) Generally conserved among land plants, more variable among algae Genes for plastid gene expression rRNAs, tRNAs ribosomal proteins RNA polymerase Genes involved in photosynthesis 28 thylakoid proteins Photosystem I (psa) Photosystem II (psb) ATP synthase subunits (atp) NADH dehydrogenase subunits (nad) Cytochrome b6f subunits (pet) RUBISCO large subunit (rbcL) (rbcS is nuclear encoded) Plastid genomes encode integral membrane components of the photosynthetic complexes Photosynthetic composition of the thylakoid membrane Green = plastid-encoded subunits Red = nuclear-encoded subunits • What do you notice about the plastid vs nuclearencoded subunits ? • What hypotheses does this suggest regarding the reasons for a plastid genome? [Leister, Trends Genet 19:47] Plastid genome transformation DNA delivery by particle bombardment or PEG precipitation DNA incorporation by homologous recombination Initial transformants are heteroplasmic, having a mixture of transformed and non-transformed plastids Selection for resistance to spectinomycin (spec) and streptomycin (strep) antibiotics that inhibit plastid protein synthesis Spec or strep resistance conferred by individual 16S rRNA mutations Spec and strep resistance conferred by aadA gene (aminoglycoside adenylyl transferase) Untransformed callus bleached; transformed callus greens and can be regenerated Multiple selection cycles may be required to obtain homoplasmy (all plastid genomes of the same type) Plastid genome transformation [Bock & Khan, Trends Biotechnol 22:311] Selection for plastid transformants A) leaf segments post bombardment with the aadA gene B) leaf segments after selection on spectinomycin; C) transfer of transformants to spectinomycin + streptomycin D) recovery of homoplasmic spec + strep resistant transformants [Bock , J Mol Biol 312:425] Applications of plastid genome transformation by homologous recombination [Bock, Curr Opin Biotechnol 18:100] Functional analysis of plastid ycf6 in transgenic plastids [Hager et al. EMBO J 18:5834] Functional analysis of plastid ycf6 in transgenic plastids ycf6 knock-out lines: •Homoplasmic for aadA insertion into ycf6 •Pale-yellow phenotype •Normal PSI function and subunit accumulation •Normal PSII function and subunit accumulation •Abnormal b6f (PET) subunit accumulation •Mass spectrometry demonstrates YCF6 in normal plastid PET complex Why, if ycf6 is the disrupted gene, does another PET complex subunit (PETA) fail to accumulate ? [Hager et al. EMBO J 18:5834] Non-Functional DNA transfer from organelle to nuclear genome Frequent Continual (can detect in “real-time” as well as evolutionary time) In large pieces e.g. Arabidopsis 262 kb numtDNA (nuclearlocalized mitochondrial DNA) 88,000 years ago e.g. Rice 131 kb nupDNA (nuclear-localized plastid DNA) 148,000 years ago Non-functional plastid-to-nucleus DNA transfer • Transform plastids with: plastid promoter – aadA linked to nuclear promoter - neo • Pollinate wild-type plants with transformants • % seed germination on kanamycin ~ frequency of nuclear promoter - neo transferred from plastid to nucleus Why does this experiment primarily estimate the frequency of DNA transfer from plastid to nucleus, rather than the frequency of functional gene transfer from plastid to nucleus? How would you re-design the experiment to test for features of a functional gene transfer? [Timmis et al. Nat Rev Genet 5:123] Land Plant Mitochondrial Genome Organization 208-2400 kb depending on species Relatively constant coding but highly variable organization among and even within a species Physical mapping with overlapping cosmid clones •Entire complexity maps as a single “master circle” •All angiosperms except Brassica hirta have one or more recombination repeats. •Repeats not conserved among species •Direct and/or inverted orientations on the “master” •Recombination generated inversions (inverted repeats) •Recombination generated subgenomic molecules (deletions) (direct repeats), some present at very low copy number (sublimons) •Leads to complex multipartite structures Recombination across direct repeats leads to deletions (subgenomic molecules) a c b d PmeI Not I AscI c’ b’ d’ c’ b a c Pac I d b’ c’ d’ Pac I Not I a AscI b’ a’ b c’ d’ AscI Pac I c d PmeI How can these deletion (subgenomic) isomers be detected? Arabidopsis mitochondrial genome organization > > > > > Two pairs of repeats active in recombination •One direct (magenta, top left) •One inverted (blue, top left) Recombining the inverted (blue pair) creates an inversion •What has happened to the orientation of the magenta repeats (top right)? [modified from Backert et al. Trends Plant Sci 2:478] Branched rosette and linear molecules from C. album mitochondria (Backert and Börner, Curr Genet 37:304) Structural plasticity of plant mitochondrial DNA [Backert et al. Trends Plant Sci 2:478] Structural plasticity of plant organelle genomes Plastid genomes map as a single circle • Inversion isomers • Indicate recombination through the LIR Plant mitochondrial genomes map as a single master circle plus •Many subgenomic circles • Inversion isomers • Imply recombination through multiple direct & inverted repeat pairs Direct visualization via EM or FISH • Rosette/knotted/branched structures • Longer-than genome linear molecules • Shorter-than genome linear and circular molecules •Sigma molecules •Branched linear molecules •Few if any genome-length circular molecules (mitochondria only) Circular maps – linear molecules A Z Y B C X D In a circular molecule or map, fragment A is linked to B, B to C, C to D, D to X, X to Y, Y to Z and Z to A. But these linkages also hold true for linear molecules fixed terminal redundancy (e.g. phage T7) ABCDEF______________XYZABC circularly permuted monomers ABCDEF______________XYZ BCDEF______________XYZA CDEF _____________ XYZAB circularly permuted monomers & terminal redundancy (e.g. phage T4) CDEF______________XYZABCDEF DEFG____________ XYZABCDEFG EFGH___________XYZABCDEFGH linear dimers or higher multimers ABCDEF__________XYZABCDEF_________XYZ Physical structures of DNA obtained via rolling circle DNA replication [Freifelder, 1983, Molecular Biology] Recombination initiated DNA replication [Kreuzer et al. J Bacteriol 177:6844] Possible origins of structural plasticity in plant organelle genomes Complex rosette/knotted structures • nucleoids Longer-than genome linear molecules • rolling circle replication • intermolecular recombination of linear molecules Shorter-than genome linear and circular molecules • intramolecular recombination between direct repeats Sigma molecules • rolling circles • recombination of circular & linear molecules Branched linear molecules • recombination • recombination-mediated replication Few if any genome-length circular molecules • limited number of circular rolling circle replication templates Plant mitochondrial genome coding content In organello protein synthesis estimates 30-50 proteins encoded by plant mitochondrial genomes Complete sequence of A. thaliana mit genome 57 genes respiratory complex components rRNAs, tRNAs, ribosomal proteins cytochrome c biogenesis Plant mit genomes lack a complete set of tRNAs mit encoded tRNAs of mit origin mit encoded tRNAs functional transfer from the plastid genome nuclear encoded tRNAs imported into mitochondria to complete the set 42 orfs that might be genes Gene density (1 gene per 8 kb) lower than the nuclear gene density (1 gene per 4-5 kb)! Plant mitochondrial genome coding content Table 3 General features of mtDNA of angiosperms Feature Ntaa Ath Bna Bvu Osa MC (bp) 430,597 366,924 221,853 368,799 490,520 A+T content (%) 55.0 55.2 54.8 56.1 56.2 Long repeated (bp) b 34,532 11,372 2,427 32,489 127,600 Uniquec Codingd 37,549 (10.6%) 38,065 (17.3%) 34,499 (10.3%) 40,065 (11.1%) Cis-splicing introns 25,617 (6.5%) 28,312 (8.0%) 28,332 (12.9%) 18,727 (5.6%) 26,238 (7.2%) ORFse 46,773 (11.8%) 37,071 (10.4%) 20,085 (9.2%) 54,288 (16.1%) 12,009 (3.3%) cp-derived (bp) 9,942 (2.5%) 3,958 (1.1%) 7,950 g (3.6%) 2.1% h 22,593 (6.2%) Others 274,527 248,662 124,994 (69.3%) (69.9%) (57%) 65.9% 262,015 (72.2%) Gene contentf 60 52 56 39,206 (9.9%) 55 53 (from Sugiyama et al. Mol Gen Gen 272:603) Mitochondrial genomes encode integral membrane components of the respiratory complexes NAD(P)H DH external H + UQH2 **** *** inner membrane CYC H + H + III I UQ * * NAD+ NADH ATP Synthase * *** 2H2O IIII TCA cycle **** IV O2 AOX NAD(P)H DH internal intermembrane space 2H2O O2 H + ADP * matrix ATP = one mitochondria-encoded subunit There is some species-to-species variation with respect to the presence or absence of genes encoding respiratory chain subunits. What is the likely explanation for this observation? (Modified from Rasmusson et al. Annu Rev Plant Biol 55:23)