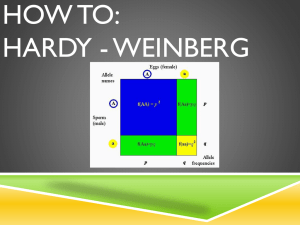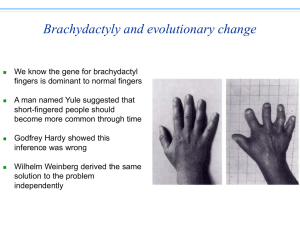
Hardy Weinberg Equilibrium
Gregor Mendel
(1822-1884)
Wilhem Weinberg
G. H. Hardy
(1877 - 1947)
(1862 – 1937)
Recall from Previous Lectures
Darwin’s Observation
Evolution acts through changes in
allele frequency at each
generation
Leads to average change in
characteristic of the population
Lectures 4-11: Mechanisms of Evolution
(Microevolution)
•
•
•
•
•
•
Hardy Weinberg Principle (Mendelian Inheritance)
Genetic Drift
Mutation
Recombination
Epigenetic Inheritance
Natural Selection
These are mechanisms acting WITHIN populations,
hence called “population genetics”—EXCEPT for
epigenetic modifications, which act on individuals
in a Lamarckian manner
4 Major Evolutionary Mechanisms
acting at the population level,
changing allele frequencies:
•
•
•
•
Genetic Drift
Natural Selection
Mutation
Migration
Testing for Hardy-Weinberg equilibrium can
be used to assess whether a population is
evolving
The Hardy-Weinberg Principle
• A population that is not evolving shows allele and
genotypic frequencies that are in Hardy Weinberg
equilibrium
• If a population is not in Hardy-Weinberg
equilibrium, it can be concluded that the
population is evolving
Fig. 23-5a
MAP
AREA
•What is a “population?”
A group of individuals within
a species that is capable of
interbreeding and producing
fertile offspring
Beaufort Sea
Porcupine
herd range
(definition for sexual species)
Fortymile
herd range
Fig. 23-5
Porcupine herd
MAP
AREA
Beaufort Sea
Porcupine
herd range
Fortymile
herd range
Fortymile herd
Hardy-Weinberg Principle
• Mathematical description of Mendelian
inheritance
Hardy-Weinberg Equilibrium
• According to the Hardy-Weinberg principle,
frequencies of alleles and genotypes in a
population remain constant from generation to
generation
• In a given population where gametes contribute
to the next generation randomly, allele
frequencies will not change
• Allelic and genotypic frequencies follow the
transmission rules of Mendelian inheritance,
which maintains constant proportions in a
population across generations
In the absence of Evolution…
Patterns of inheritance should always be in
“Hardy Weinberg Equilibrium”
Following the transmission rules of Mendel
Requirements of HW
Violation
Evolution
Large population size
Genetic drift
Random Mating
Inbreeding & other
No Mutations
Mutations
No Natural Selection
Natural Selection
No Migration
Migration
An evolving population is one that
violates Hardy-Weinberg Assumptions
“Null Model”
• No Evolution: Null Model to test if no
evolution is happening should simply be a
population in Hardy-Weinberg Equilibrium
• No Selection: Null Model to test whether
Natural Selection is occurring should have no
selection, but should include Genetic Drift
– This is because Genetic Drift is operating even
when there is no Natural Selection
Hardy-Weinberg Theorem
In a non-evolving population,
frequency of alleles and genotypes
remain constant over generations
important concepts
• gene:
A region of genome sequence (DNA or RNA), that is
the unit of inheritance , the product of which contributes to
phenotype
• locus:
Location in a genome (used interchangeably with
“gene,” if the location is at a gene… but, locus can be anywhere,
so meaning is broader than gene)
• loci: Plural of locus
• allele: Variant forms of a gene (e.g. alleles for different eye
colors, BRCA1 breast cancer allele, etc.)
• genotype: The combination of alleles at a locus (gene)
• phenotype: The expression of a trait, as a result of the
genotype and regulation of genes (green eyes, brown hair, body
size, finger length, cystic fibrosis, etc.)
important concepts
• allele: Variant forms of a gene (e.g. alleles for different eye
colors, BRCA1 breast cancer allele, etc.)
• We are diploid (2 chromosomes), so we have 2 alleles
at a locus (any location in the genome)
• However, there can be many alleles at a locus in a
population.
– For example, you might have inherited a blue eye allele from
your mom and a brown eye allele from your dad… you can’t
have more alleles than that (only 2 chromosomes, one from
each parent)
– BUT, there could be many alleles at this locus in the
population, blue, green, grey, brown, etc.
• Alleles in a
population of
diploid organisms
A2
Eggs
A1
A3
A1
A2
A1
A4
A2
Sperm
A1
A3
A4
A1
A1
Random Mating (Sex)
Zygotes
A1A1
A1A3
• Genotypes
A1A1
A1A1
A2A4
A3A1
A2
Eggs
A1
A3
A1
A2
A1
A4
So then can we
predict the % of
alleles and genotypes
in the population at
each generation?
A2
Sperm
A1
A3
A4
A1
Zygotes
A1
A1A1
A1A3
A1A1
A1A1
A2A4
A3A1
Hardy-Weinberg Theorem
In a non-evolving population,
frequency of alleles and genotypes
remain constant over generations
Fig. 23-6
Alleles in the population
Frequencies of alleles
p = frequency of
CR allele
= 0.8
q = frequency of
CW allele
= 0.2
Gametes produced
Each egg:
80%
chance
20%
chance
Each sperm:
80%
chance
Hardy-Weinberg proportions indicate the
expected allele and genotype frequencies,
given the starting frequencies
20%
chance
• By convention, if there are 2 alleles at a locus, p and
q are used to represent their frequencies
• The frequency of all alleles in a population will add
up to 1
– For example, p + q = 1
If p and q represent the relative frequencies of the
only two possible alleles in a population at a
particular locus, then for a diploid organism (2
chromosomes),
(p + q) 2 = 1
= p2 + 2pq + q2 = 1
– where p2 and q2 represent the frequencies of the
homozygous genotypes and 2pq represents the
frequency of the heterozygous genotype
What about for a triploid organism?
What about for a triploid organism?
• (p + q)3 = 1
= p3 + 3p2q + 3pq2 + q3 = 1
Potential offspring: ppp, ppq, pqp, qpp,
qqp, pqq, qpq, qqq
How about tetraploid? You work it out.
Hardy Weinberg Theorem
ALLELES
Probability of A = p
Probability of a = q
p+q=1
GENOTYPES
AA: p x p =
p2
Aa: p x q + q x p = 2pq
aa: q x q =
q2
p2 + 2pq + q2 = 1
More General HW Equations
• One locus three alleles: (p + q + r)2 = p2 + q2 + r2 + 2pq +2pr +
2qr
• One locus n # alleles: (p1 + p2 + p3 + p4 … …+ pn)2 = p12 + p22 +
p32 + p42… …+ pn2 + 2p1p2 + 2p1p3 + 2p2p3 + 2p1p4 + 2p1p5 + …
… + 2pn-1pn
• For a polyploid (more than two chromosomes):
(p + q)c, where c = number of chromosomes
• If multiple loci (genes) code for a trait, each locus follows the
HW principle independently, and then the alleles at each loci
interact to influence the trait
ALLELE Frequencies
Frequency of A = p = 0.8
Frequency of a = q = 0.2
p+q=1
Expected GENOTYPE Frequencies
AA: p x p = p2 = 0.8 x 0.8 = 0.64
Aa: p x q + q x p = 2pq
= 2 x (0.8 x 0.2) = 0.32
aa: q x q = q2 = 0.2 x 0.2 = 0.04
Allele frequencies remain the same at
next generation
p2 + 2pq + q2
= 0.64 + 0.32 + 0.04 = 1
Expected Allele Frequencies at 2nd Generation
p = AA + Aa/2 = 0.64 + (0.32/2) = 0.8
q = aa + Aa/2 = 0.04 + (0.32/2) = 0.2
Hardy Weinberg Theorem
ALLELE Frequency
Frequency of A = p = 0.8
Frequency of a = q = 0.2
p+q=1
Expected GENOTYPE Frequency
AA:
Aa:
aa :
pxp=
p2 = 0.8 x 0.8 = 0.64
p x q + q x p = 2pq = 2 x (0.8 x 0.2) = 0.32
qxq=
q2 = 0.2 x 0.2 = 0.04
p2 + 2pq + q2 = 0.64 + 0.32 + 0.04 = 1
Expected Allele Frequency at 2nd Generation
p = AA + Aa/2 = 0.64 + (0.32/2) = 0.8
q = aa + Aa/2 = 0.04 + (0.32/2) = 0.2
Similar example,
But with different starting allele frequencies
p
q
p2
2pq
q2
Fig. 23-7-4
80% CR ( p = 0.8)
20% CW (q = 0.2)
Sperm
CR
(80%)
64% ( p2)
CR CR
16% (qp)
CR CW
CW
(20%)
16% ( pq)
CR CW
4% (q2)
CW CW
64% CR CR, 32% CR CW, and 4% CW CW
Gametes of this generation:
64% CR + 16% CR = 80% CR = 0.8 = p
4% CW
+ 16% CW
= 20% CW = 0.2 = q
Genotypes in the next generation:
64% CR CR, 32% CR CW, and 4% CW CW plants
Perform the same
calculations using
percentages
Fig. 23-7-1
80% CR (p = 0.8)
20% CW (q = 0.2)
Sperm
CR
(80%)
CW
(20%)
64% (p2)
CRCR
16% (pq)
CRCW
16% (qp)
CRCW
4% (q2)
CW CW
Fig. 23-7-2
64% CRCR, 32% CRCW, and 4% CWCW
Gametes of this generation:
64% CR + 16% CR
= 80% CR = 0.8 = p
4% CW + 16% CW = 20% CW = 0.2 = q
Fig. 23-7-3
64% CRCR, 32% CRCW, and 4% CWCW
Gametes of this generation:
64% CR + 16% CR
= 80% CR = 0.8 = p
4% CW + 16% CW = 20% CW = 0.2 = q
Genotypes in the next generation:
64% CRCR, 32% CRCW, and 4% CWCW plants
Calculating Allele Frequencies from # of Individuals
• The frequency of an allele in a population can be
calculated from # of individuals:
– For diploid organisms, the total number of alleles at
a locus is the total number of individuals x 2
– The total number of dominant alleles at a locus is 2
alleles for each homozygous dominant individual
– plus 1 allele for each heterozygous individual; the
same logic applies for recessive alleles
Calculating Allele and Genotype Frequencies from
# of Individuals
AA
120
Aa
60
aa
35 (# of individuals)
#A = (2 x AA) + Aa = 240 + 60 = 300
#a = (2 x aa) + Aa = 70 + 60 = 130
Proportion A = 300/total = 300/430 = 0.70
Proportion a = 130/total = 130/430 = 0.30
A + a = 0.70 + 0.30 = 1
Proportion AA = 120/215 = 0.56
Proportion Aa = 60/215 = 0.28
Proportion aa = 35/215 = 0.16
AA + Aa + aa = 0.56 + 0.28 +0.16 = 1
Applying the Hardy-Weinberg Principle
• Example: estimate frequency of a disease allele in
a population
• Phenylketonuria (PKU) is a metabolic disorder that
results from homozygosity for a recessive allele
• Individuals that are homozygous for the deleterious
recessive allele cannot break down phenylalanine, results
in build up mental retardation
• The occurrence of PKU is 1 per 10,000 births
• How many carriers of this disease in the
population?
– Rare deleterious recessives often remain in a
population because they are hidden in the
heterozygous state (the “carriers”)
– Natural selection can only act on the homozygous
individuals where the phenotype is exposed
(individuals who show symptoms of PKU)
– We can assume HW equilibrium if:
• There is no migration from a population with different
allele frequency
• Random mating
• No genetic drift
• Etc
So, let’s calculate HW frequencies
• The occurrence of PKU is 1 per 10,000 births
(frequency of the disease allele):
q2 = 0.0001
q = sqrt(q2 ) = sqrt(0.0001) = 0.01
• The frequency of normal alleles is:
p = 1 – q = 1 – 0.01 = 0.99
• The frequency of carriers (heterozygotes) of the
deleterious allele is:
2pq = 2 x 0.99 x 0.01 = 0.0198
or approximately 2% of the U.S. population
Conditions for Hardy-Weinberg Equilibrium
• The Hardy-Weinberg theorem describes a
hypothetical population
• The five conditions for nonevolving populations
are rarely met in nature:
–
–
–
–
–
No mutations
Random mating
No natural selection
Extremely large population size
No gene flow
• So, in real populations, allele and genotype
frequencies do change over time
DEVIATION
from
Hardy-Weinberg Equilibrium
Indicates that
EVOLUTION
Is happening
4 Major Evolutionary Mechanisms:
•
•
•
•
Genetic Drift
Natural Selection
Mutation
Migration
Hardy-Weinberg across a Genome
• In natural populations, some loci might be out of
HW equilibrium, while being in Hardy-Weinberg
equilibrium at other loci
• For example, some loci might be undergoing
natural selection and become out of HW
equilibrium, while the rest of the genome remains
in HW equilibrium
Allele A1 Demo
Examples of Deviation from
Hardy-Weinberg Equilibrium
What would Genetic Drift look
like?
• Most populations are experiencing some level
of genetic drift, unless they are incredibly
large
Examples of Deviation from
Hardy-Weinberg Equilibrium
Generation 1
Generation 2
Generation 3
Generation 4
AA
0.64
0.63
0.64
0.65
Aa
0.32
0.33
0.315
0.31
aa
0.04
0.04
0.045
0.04
Is this population in HW equilibrium?
If not, how does it deviate?
What could be the reason?
Examples of Deviation from
Hardy-Weinberg Equilibrium
Generation 1
Generation 2
Generation 3
Generation 4
AA
0.64
0.63
0.64
0.65
Aa
0.32
0.33
0.315
0.31
aa
0.04
0.04
0.045
0.04
This is a case of Genetic Drift, where
allele frequencies are fluctuating
randomly across generations
Examples of Deviation from
Hardy-Weinberg Equilibrium
AA
0.64
Aa
0.32
aa
0
Is this population in HW equilibrium?
If not, how does it deviate?
What could be the reason?
Examples of Deviation from
Hardy-Weinberg Equilibrium
AA
0.64
Aa
0.32
aa
0
Here this appears to be Directional
Selection favoring AA
Or… Negative Selection disfavoring aa
Examples of Deviation from
Hardy-Weinberg Equilibrium
AA
0.25
Aa
0.70
aa
0.05
Is this population in HW equilibrium?
If not, how does it deviate?
What could be the reason?
Examples of Deviation from
Hardy-Weinberg Equilibrium
AA
0.25
Aa
0.70
aa
0.05
This appears to be a case of Heterozygote
Advantage (or Overdominance)
Examples of Deviation from
Hardy-Weinberg Equilibrium
AA
0.10
Aa
0.10
aa
0.80
Is this population in HW equilibrium?
If not, how does it deviate?
What could be the reason?
Examples of Deviation from
Hardy-Weinberg Equilibrium
AA
0.10
Aa
0.10
aa
0.80
Selection appears to be favoring aa
How can you tell whether a population
is out of HW Equilibrium?
Example: Does this population remain in
Hardy Weinberg Equilibrium across
Generations?
Generation 1
Generation 2
Generation 3
AA
0.25
0.20
0.10
Aa
0.50
0.60
0.80
aa
0.25
0.20
0.10
Generation 1
Generation 2
Generation 3
AA
0.25
0.20
0.10
Aa
0.50
0.60
0.80
aa
0.25
0.20
0.10
In this case, allele frequencies (of A and a) did not
change.
***However, the population did go out of HW
equilibrium because you can no longer predict
genotypic frequencies from allele frequencies
For example, p = 0.5, p2 = 0.25, but in Generation 3,
the observe p2 = 0.10
How can you tell whether a population
is out of HW Equilibrium?
1. When allele frequencies are changing across
generations
2. When you cannot predict genotype
frequencies from allele frequencies (means
there is an excess or deficit of genotypes
than what would be expected given the allele
frequencies)
• One generation of Random Mating could put a
population back into Hardy Weinberg
Equilibrium
Summary
(1) A nonevolving population is in HW
Equilibrium
(2) Evolution occurs when the requirements for
HW Equilibrium are not met
(3) HW Equilibrium is violated when there is
Genetic Drift, Migration, Mutations, Natural
Selection, and Nonrandom Mating
Hardy Weinberg Equilibrium
Gregor Mendel
(1822-1884)
Wilhem Weinberg
G. H. Hardy
(1877 - 1947)
(1862 – 1937)
1. Nabila is a Saudi Princess who is arranged to marry her
first cousin. Many in her family have died of a rare blood
disease, which sometimes skips generations, and thus
appears to be recessive. Nabila thinks that she is a carrier of
this disease. If her fiancé is also a carrier, what is the
probability that her offspring will have (be afflicted with) the
disease?
(A) 1/4
(B) 1/3
(C) 1/2
(D) 3/4
(E) zero
The following are numbers of pink and white flowers in a population.
Generation 1:
Generation 2:
Generation 3:
Pink
901
1204
1510
White
302
403
504
2. Which of the following is most likely to be TRUE?
(A) The heterozygotes are probably pink
(B) The recessive allele here (probably white) is clearly deleterious
(C) Evolution is occurring, as allele frequencies are changing greatly over time
(D) Clearly there is a heterozygote advantage
(E) The frequencies above violate Hardy-Weinberg expectations
The following are numbers of purple and white peas in a population.
Generation 1:
Generation 2:
Generation 3:
(A1A1)
Purple
360
100
0
(A1A2)
Purple
480
200
100
(A2A2)
White
160
200
300
3. What are the genotype frequencies at each generation?
(A) Generation 1: 0.30, 0.50, 0.20
Generation 2: 0.20, 0.40, 0.40
Generation 3: 0, 0.333, 0.666
(B) Generation 1: 0.36, 0.48, 0.16
Generation 2: 0.10, 0.20, 0.20
Generation 3: 0, 0.10, 0.30
(C) Generation 1: 0.36, 0.48, 0.16
Generation 2: 0.20, 0.40, 0.40
Generation 3: 0, 0.25, 0.75
(D) Generation 1: 0.36, 0.48, 0.16
Generation 2: 0.36, 0.48, 0.16
Generation 3: 0.36, 0.48, 0.16
4. From the example on the previous slide, what are the
frequencies of alleles at each generation?
(A) Generation1: Dominant allele (A1) = 0.6, Recessive allele (A2) = 0.4
Generation2: Dominant allele = 0.4, Recessive allele = 0.6
Generation3: Dominant allele = 0.125, Recessive allele = 0.875
(B) Generation1: Dominant allele = 0.6, Recessive allele = 0.4
Generation2: Dominant allele = 0.6, Recessive allele = 0.4
Generation3: Dominant allele = 0.6, Recessive allele = 0.4
(C) Generation1: Dominant allele = 0.6, Recessive allele = 0.4
Generation2: Dominant allele = 0.5, Recessive allele = 0.5
Generation3: Dominant allele = 0.25, Recessive allele = 0.75
(D) Generation1: Dominant allele = 0.4, Recessive allele = 0.6
Generation2: Dominant allele = 0.5, Recessive allele = 0.5
Generation3: Dominant allele = 0.25, Recessive allele = 0.75
5. From the example two slides ago, which evolutionary
mechanism might be operating across generations?
(A) Mutation
(B) Selection favoring A1
(C) Heterozygote advantage
(D) Selection favoring A2
(E) Inbreeding
Answers:
1. Parents: Aa x Aa = Offspring: AA (25%), Aa (50%), aa (25%)
Answer = A
2. A
3. C
4. A
5. D