Transformation Lab

TRANSFORMATION
Intro to Lab #8
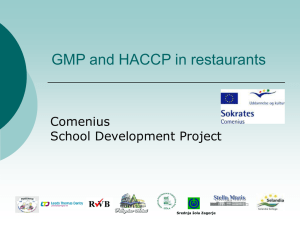
Figure 20.2
Bacterium
1 Gene inserted into plasmid
Cell containing gene of interest
Bacterial chromosome
Plasmid
Recombinant
DNA (plasmid)
Gene of interest
2 Plasmid put into bacterial cell
DNA of chromosome
(
“foreign” DNA)
Recombinant bacterium
Gene of interest
Copies of gene
3 Host cell grown in culture to form a clone of cells containing the “cloned” gene of interest
Protein expressed from gene of interest
Protein harvested
Basic research on gene
4 Basic research and various applications
Basic research on protein
Gene for pest resistance inserted into plants
Gene used to alter bacteria for cleaning up toxic waste
Protein dissolves blood clots in heart attack therapy
Human growth hormone treats stunted growth
1.
2.
3.
4.
5.
6.
7.
8.
Transformation
Identify a restriction enzyme that will cut out gene of interest and cut open the plasmid
Isolate DNA from 2 sources (making sure the at the plasmid has a genetic marker)
Cut both types of DNA with the same restriction enzyme
Mix the 2 types of DNA to join them through complementary base pairing
Add DNA ligase to bond DNA covalently producing Recombinant DNA
Incubate bacteria at 42 C with calcium chloride; bacteria become competent / permeable - so that the bacteria will take in the plasmid
(TRANSFORMATION)
Use a genetic marker to identify bacteria with the recombinant plasmid
Clone bacteria
Pre-Lab Inquiry
You work on a team in a research lab. You have three separate plasmids in your lab, but the labels have come off of the tubes and you no longer know which plasmid is which. For the time being, you will refer to the plasmids are plasmids 1, 2, and 3. You know the following:
One plasmid has kanamycin-resistant gene.
Two plasmids have an ampicillin-resistance gene.
In addition to having an antibiotic-resistance marker, one plasmid also codes for the gene for green fluorescent protein (GFP), a protein from the bioluminescent jellyfish Aequorea victoria.
Pre-lab Inquiry
Bacteria that produce the green fluorescent protein look green under white light and fluoresce under ultraviolet light
Assume that each research team has access to the following materials and is responsible for identifying one of the three plasmid. (Note: you will not actually use any materials during this pre-lab inquiry). Before planning your experiment, answer the following questions to ensure that you understand some of the basic concepts involved in the lab. Some of these questions are specifically designed to help you think about setting up your experiment.
Materials Available
Starter plates (containing E. coli in rapid growth)
Kanamycin plates
Ampicillin plates
LB plates (plates contain Luria Broth/no antibiotic)
Plasmids 1, 2, and 3
Transformation agents, tubes, and equipment
(inoculating loops, calcium chloride, ice, 42C water bath, incubator)
Questions
1. What is a plasmid?
Small, usually circular, extra-chromosomal piece of
DNA that exits in nature in some bacteria and yeasts.
They can be transferred between organisms. In the lab they can be used to manipulate and introduce DNA of interest into bacterium.
2. What is transformation?
The uptake of exongenous, naked DNA by a cell. The newly adopted DNA can become a heritable part of the cell’s genetic material.
Questions
3. Why is naturally occurring transformation beneficial to bacteria?
It provides them with access to new genetic material, which increases their chances of being able to adapt to the environment.
4. Why is transformation useful to research scientists?
It enables them to introduce foreign DNA into bacteria.
This allows them to further study and work with genes on the plasmid DNA and the proteins that the genes code for.
Questions
5. Should you plate some of your transformed bacteria onto plates with antibiotic? Why or Why not?
Yes – you will select for/isolate bacteria that have been transformed with the plasmid with the resistance gene
6. What would you expect to see if you plated some of your transformed bacteria onto a plate without antibiotic? Would there be an advantage to doing this?
Explain.
You would see a lawn of bacteria (the plate would be covered). This tells you that the bacteria survived the transformation process. When compared to the antibiotic plates it gives an indication of how many bacteria were transformed
Questions
7. To transform bacteria with plasmids, technicians first make the bacteria competent (capable of taking up
DNA) by placing them in calcium chloride and chilling them. Plasmid is then added to the competent bacteria and the plasmid/bacteria combination is taken through a few more steps to make the bacteria take up DNA.
In your experiment, should you treat a tube of bacteria that you do not add plasmid to exactly as you do the tube of bacteria that you will transform? Why or why not?
This provides a control demonstrating that the antibioticresistance colonies that appear on the plate are a result of the plasmid being taken up by the cells. If no plasmid, then there should be no growth on antibiotic plates.
What would you expect?
LB AMP plates LB Kan plates LB plates plasmid + plmd - plmd + plmd - plmd + plmd - plmd Color when present
Amp
Kan
Amp & GFP
What would you expect?
LB AMP plates LB Kan plates LB plates plasmid + plmd - plmd + plmd - plmd + plmd - plmd Color when present
Amp 25 -100 colonies
0 colonies
0 colonies
0 colonies
Lawn Lawn yellow
Kan Lawn Lawn Yellow
Amp & GFP
0 colonies
25-100 colonies
0 colonies
0 colonies
25-100 colonies
0 colonies
0 colonies
0 colonies
Lawn Lawn Yellow/gre en or green
What you need…
2 LB plates
2 LB/Kan plates
2 LB/Amp plates
2 sterile transformation tubes & rack
Container with crushed ice
3 sterile inoculating loops
6 sterile transfer pipets
Waste container
3 mL vial of LB
3 mL vial of CaCl
2
(on ice)
Basic Outline of the Lab
1.
2.
3.
4.
5.
6.
7.
8.
Label 1 tube + / 1 tube –
Add CaCl
2
, place on ice
Use sterile loop to transfer E. coli to both tubes (no agar!) return to ice
Use sterile loop to add loopful of plasmid (1, 2, or 3) to + tube
Ice for 15 min & label 1of each plate “+” and 1 of each “-”
Heat shock (42C for 90 sec)
Add luria broth & sit at room temp
5 to 15 minutes
Spread bacteria on plates