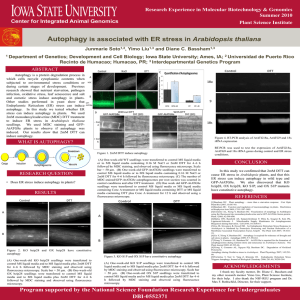
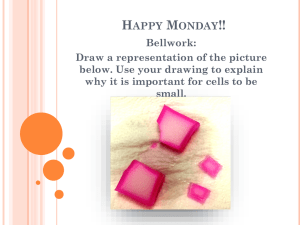
Apoptosis Autophagy
advertisement

Apoptosis Classification of cell death Cell death Physiological Necrotic apoptosis Caspase-dependent receptor-caspase 8 autophagic other Caspase-independent mitochondria-caspase 9 Apoptosis • Apoptosis (1972) – Greek word “falling off” • Built-in (programmed) mechanism) • or self-destructionsuicide • Type of programmed cell death based upon morphological features Apoptosis • Apoptosis is programmed cell death, which prevents damage to neighboring cells by controlling how the affected cell dies • During apoptosis, the cell's cytoskeleton is broken down, causing multiple bulges in the membrane. This is called blebbing. • These blebs (a.k.a. apoptotic bodies) can separate from the cell, taking a portion of cytoplasm with them. Phagocytic cells eventually consume these fragments. • Hence, apoptosis keeps damaged cellular contents from spilling out and damaging other cells. Two lymphocytes Healthy Staurosporin-treated HeLa cells Apoptosi s Apoptosi s Apoptoti c Weinberg Fig 9.18 Different parts of the apoptotic programme. Markers for the process. Fragmentation of DNA Pyknotic nuclear fragments Apoptotic cell Phagocytosis of apoptotic bodies Fragmentation of Golgi bodies Phosphorylation of Histone 2B APOPTOSIS SIGNALS Mitochondriadependent apoptosis Caspasedependent apoptosis Caspaseindependent apoptosis Nature Reviews Cancer 2; 647-656 (2002) Death Receptordependent apoptosis Caspasedependent apoptosis Caspaseindependent necrosis Physiological Relevance of Apoptosis • • • • • • • • • • Embryonic Development Regulation of/by Immune System Negative Selection CTL Killing (eg. Immune surveillance, viral infections) Terminating Active Immune Response Tight Regulation of Cell Number (eg. BM, GI, Uterus, Skin) Compensatory Response to Cell Stress Intrinsic Pathway (e.g. GF removal, XRT, Chemo.) Extrinsic Pathway (e.g. FAS.Ag, TNF-R Activation) Senescence (ageing) Necrosis • Necrosis is traumatic cell death caused by injury. Necrosis of cells will lyse and damage neighboring cells by spilling all the intracellular contents. This causes inflammation of neighboring tissues and further trauma. Morphologic appearance of necrosis • Increased eosinophilia: Loss of RNA in the cytoplasm Increased binding of eosin to denatured cytoplasmic protein • More glassy homogeneous appearance Loss of glycogen particles • Vacuolated and moth-eaten cytoplasm • Calcification of necrotic cells Principle Sites of Damage in Cell Injury Necrosis vs. Apoptosis • Apoptosis – programmed cell death • Necrosis – un-programmed cell death Necrotic cells Necrotic cell with multiple lesions Apoptotic cell with blebbing Contrast of Apoptosis and necrosis Apoptosis Necrosis Death by apoptosis is a neat, orderly process The Roles of Therapy-Induced Autophagy and Necrosis in Cancer Treatment • Inhibiting autophagy for cancer therapy. A, surgery, chemotherapy, targeted therapies, and radiation can all activate autophagy. Treatment of cancer cells with chloroquine (CQ) derivatives leads to deacidification of lysosomes followed by an accumulation of ineffective autophagic vesicles. In cells dependent on autophagy for survival, autophagy inhibition with chloroquine leads to cell death. B and C, electron micrographs of PC3 prostate cancer cells either untreated (B) or treated with chloroquine (C). Bar, 2 μm. The Roles of Therapy-Induced Autophagy and Necrosis in Cancer Treatment • • Regulation of autophagy and necrosis. A, autophagy. Multiple stresses, including interrupted growth factor signaling, activate autophagy. Fluctuations in small molecules, such as increased ROS, may be the link between signal transduction events and the enzymatic activation of autophagy genes. Once initiated, autophagic vesicles form around damaged mitochondria and proteins. This double-membrane structure fuses with the lysosome resulting in the degradation of contents. PI3K, phosphatidylinositol 3-kinase; mTOR, mammalian target of rapamycin. B, necrosis. Initiators of necrosis include the NAD+depleting PARP and the death receptor adaptors RIPK1 and TRAF2. Activated RIPK1 translocates to the mitochondria, disrupting the association of cyclophilin D (Cyc. D) and adenosine nucleotide translocator (ANT). The subsequent depletion of NAD+ and ATP and accumulation of calcium and ROS lead to activation of calpain proteases and lipid peroxidation followed by widespread membrane disruption. Clin Cancer Res 2007;13:7271-7279. The Roles of Therapy-Induced Autophagy and Necrosis in Cancer Treatment • • • • • • • • Mechanistic interplay between apoptosis and autophagy Research in the Lane Lab focuses of mechanisms of cell death, with a particular interest in the molecular relationships between apoptosis and autophagy. Autophagy is activated during many forms of cell stress, and is upregulated during terminal differentiation. It is generally regarded as a protective mechanism in cells undergoing starvation and oxidative stress, but has been proposed to switch to a cell death mechanism in its own right when other death pathways (such as apoptosis) are prevented. Evidence suggests that apoptosis and autophagy are linked by common regulatory factors (see below), so identifying and characterising molecules and pathways that couple apoptosis and autophagy may lead to novel therapeutic mechanisms for cancer and neurodegeneration - important diseases that feature both of these cellular processes. Apoptosis Autophagy - Molecules identified todate that play dual roles in apoptosis and autophagy include Beclin-1 (a Bcl-2/BclXL interactor that forms the core of a multi-component, type III PI3-kinase involved in autophagosome membrane supply) and Atg5 (a factor required for autophagosomal membrane biogenesis and Atg8 recruitment which is cleaved by calpains becoming highly cytotoxic). We have reported that Atg4D (a member of the Atg4 endopeptidase family required for Atg8 processing) is cleaved by caspases, whereupon it gains Atg8 processing activity but acquires apoptosis-inducing capabilities. Autophagy (macroautophagy) is characterised by the formation of double membrane-bound organelles that sequester regions of cytoplasm including misfolded protein aggregates and organelles.The autophagosome membrane is decorated with a protein known as Atg8 (mammalian orthologues include LC3, Gate-16 and GABARAP), enabling quantitation of autophagy upregulation by fluorescence microscopy of Atg8 puncta combined with automated microscopy. EM is also routinely used to identify/quantitate autophagosomes. Light microscopy - dynamic assessment Electron microscopy - high resolution assessment • Molecules identified todate that play dual roles in apoptosis and autophagy include Beclin-1 (a Bcl-2/Bcl-XL interactor that forms the core of a multicomponent, type III PI3-kinase involved in autophagosome membrane supply) and Atg5 (a factor required for autophagosomal membrane biogenesis and Atg8 recruitment which is cleaved by calpains becoming highly cytotoxic). We have reported that Atg4D (a member of the Atg4 endopeptidase family required for Atg8 processing) is cleaved by caspases, whereupon it gains Atg8 processing activity but acquires apoptosis-inducing capabilities. Autophagy and its inhibitors From the following article: Self-eating and self-killing: crosstalk between autophagy and apoptosis M. Chiara Maiuri, Einat Zalckvar, Adi Kimchi & Guido Kroemer Nature Reviews Molecular Cell Biology 8, 741-752 (September 2007) • • • Autophagy starts with the stepwise engulfment of cytoplasmic material (cytosol and/or organelles) by the phagophore (also called isolation membrane), which sequesters material in double-membraned vesicles named autophagosomes (also called autophagic vacuoles). In many cellular settings, the first regulatory process (see figure, step 1) involves the de-repression of the mTOR Ser/Thr kinase, which inhibits autophagy by phosphorylating autophagy protein-13 (Atg13). This phosphorylation leads to the dissociation of Atg13 from a protein complex that contains Atg1 kinase and Atg17, and thus attenuates the Atg1 kinase activity. When mTOR is inhibited, re-association of dephosphorylated Atg13 with Atg1 stimulates its catalytic activity and induces autophagy. Notably, the mammalian orthologue of the yeast Atg13 has not been identified to date. Among the initial steps of vesicle nucleation is the activation of mammalian Vps34, a class III phosphatidylinositol 3-kinase (PI3K), to generate phosphatidylinositol-3-phosphate (PtdIns3P) (step 2). Vps34 activation depends on the formation of a multiprotein complex in which beclin-1 (Becn1; the mammalian orthologue of Atg6), UVRAG (UV irradiation resistance-associated tumour suppressor gene) and a myristylated kinase (Vps15, or p150 in humans) participate. Two ubiquitin-like conjugation systems are part of the vesicle elongation process (step 3). One pathway involves the covalent conjugation of Atg12 to Atg5, with the help of the E1-like enzyme Atg7 and the E2-like enzyme Atg10. The second pathway involves the conjugation of phosphatidylethanolamine (PE) to LC3/Atg8 (LC3 is one of the mammalian homologues of Atg8) by the sequential action of the protease Atg4, the E1-like enzyme Atg7 and the E2-like enzyme Atg3. Lipid conjugation leads to the conversion of the soluble form of LC3 (named LC3-I) to the autophagic-vesicle-associated form (LC3-II). LC3-II is used as a marker of autophagy because its lipidation and specific recruitment to autophagosomes provides a shift from diffuse to punctate staining of the protein and increases its electrophoretic mobility on gels compared with LC3I. Moreover, green fluorescent protein–LC3 fusion proteins can be used to visualize autophagosomes by fluorescence videomicroscopy91. The mechanism of retrieval in which the Atg9 complex participates is poorly studied (step 4). Autophagosomes undergo maturation by fusion with lysosomes to create autolysosomes (steps 5 and 6). In the autolysosomes, the inner membrane as well as the luminal content of the autophagic vacuoles is degraded by lysosomal enzymes that act optimally within this acidic compartment4, 92. Pharmacological inhibitors and small interfering RNAs that are capable of inhibiting distinct steps of this process are shown (red blocking arrows). Bcl2 and Bcl-XL are regulators of beclin-1. Lamp2, lysosome-associated membrane glycoprotein-2. Caspase-dependent and caspase-independent routes to cell death From the following article: Self-eating and self-killing: crosstalk between autophagy and apoptosis M. Chiara Maiuri, Einat Zalckvar, Adi Kimchi & Guido Kroemer Nature Reviews Molecular Cell Biology 8, 741-752 (September 2007) • Two main pathways lead to caspase-dependent apoptosis (see figure). The extrinsic pathway involves stimulation of members of the tumour necrosis factor receptor (TNFR) superfamily, such CD95/Fas, TNFR or TRAILR (death receptors). The intrinsic pathway is characterized by mitochondrial outer membrane permeabilization (MOMP) and the release of mitochondrial cytochrome c, which results in assembly of a caspase-activating complex between caspase-9 and APAF1 (the apoptosome). Death-receptor stimulation typically results in the recruitment and activation of caspase-8 by the Fas-associated via death domain (FADD)/TNFR1-associated death domain protein (TRADD) to form a death-inducing signalling complex (DISC) that can further propagate death signals in three ways: via proteolysis of the BCL2 homology-3 (BH3)-only protein BID, which provokes translocation of truncated BID to mitochondria and consequent MOMP; by direct proteolytic activation of downstream effector caspases; or via activation of the kinase RIP93. In the intrinsic pathway, a range of BH3-only proteins act as sentinels for cell stress, organellespecific damage or infection, and can be mobilized via post-translational modification (such as proteolysis or phosphorylation) or subcellular relocalization to initiate MOMP. The BH3-only proteins stimulate MOMP by triggering the oligomerization of BAX and/or BAK in the outer mitochondrial membrane, thereby forming channels that permit the escape of multiple proteins from the mitochondrial intermembrane space26, 56, 57. In the context of DNA damage, stabilization of the p53 tumour-suppressor protein can result in transcriptional activation of the BH3-only proteins (such as PUMA and NOXA) that can promote MOMP via the BAX/BAK channel52. Alternatively, DNA damage can activate caspase-2 in a multinuclear complex that involves the p53-induced protein with a death domain (PIDD) and the RIP-associated protein with a death domain (RAIDD) (together known as the piddosome)94. Caspase-2 may then induce MOMP and/or direct caspase activation. Several factors among the mitochondrial proteins that are released as a result of MOMP (apoptosis-inducing factor (AIF), Omi, EndoG) can promote caspase-independent cell death95, which can also result from stimuli that cause lysosomal membrane permeabilization (LMP), resulting in the release of cathepsin proteases into the cytosol9. Such cathepsins can also trigger MOMP, thereby stimulating the mitochondrial pathway of apoptosis. JNK, c-Jun N-terminal kinase; LMP, lysosomal membrane permeabilization; ROS, reactive oxygen species. One protein, multiple functions: against molecular reductionism From the following article: Self-eating and self-killing: crosstalk between autophagy and apoptosis M. Chiara Maiuri, Einat Zalckvar, Adi Kimchi & Guido Kroemer Nature Reviews Molecular Cell Biology 8, 741-752 • • Many cell-death researchers assumed that the sole function of a whole series of proteins — including the BCL2-family proteins, caspases and their activators — was the regulation of apoptosis. However, recent work has revealed that caspases regulate normal activation and differentiation processes96, 97 and that BCL2 proteins may regulate neural plasticity98. Even in Caenorhabditis elegans, it was found that the sole BCL2 homology-3 (BH3)-only protein EGL-1, which is required for developmental cell death, is also required for non-lethal autophagy induced by starvation58. Similarly, an essential component of the caspase activation complex, CED-4 (the nematode equivalent of APAF1), has a non-apoptotic role in the DNA-damage checkpoint99. Some of the Atg proteins, which are required for autophagy, may also have unsuspected roles outside of the autophagic process. One prominent example is ATG5, which, under certain circumstances, is proteolytically activated to become a pro-apoptotic molecule that translocates to mitochondria and triggers mitochondrial outer membrane permeabilization (MOMP)51. Another example is beclin-1, which bears a BH3 domain58, 62 that might favour apoptosis, at least in some circumstances. As a result, it may be an oversimplification to assume that the inhibition of cell death in response to DNA damage or the activation of apoptosis/necrosis during nutrient deprivation (which both result from the knockout or knockdown of a single Atg gene) is due to suppression of the autophagic process. Several genes from different modules of autophagy should be knocked down independently to find out whether the resulting phenotype is similar before final conclusions are drawn. The same holds for experiments in which apoptotic pathways have been interrupted by knocking out or knocking down single genes. A model for the roles of apoptosis and autophagy in tumorigenesis. • A common cellular response to metabolic stress is cell death mediated by apoptosis, which limits tumour growth. Tumours may trigger autophagymediated cell survival in certain metabolicstressed tumour regions. In apoptotic-defective, metabolic-stressed tumour cells, activation of autophagy prevents death from necrosis, whereas defects in autophagy lead to accumulation of p62, damaged mitochondria, ROS and protein aggregates, resulting in genome damage and tumorigenesis. For additional information, see refs 68 and 119. The relationship between apoptosis and autophagy Self-eating and self-killing: crosstalk between autophagy and apoptosis M. Chiara Maiuri, Einat Zalckvar, Adi Kimchi & Guido Kroemer Nature Reviews Molecular Cell Biology 8, 741-752 (September 2007) • Similar stressors can induce either apoptosis or autophagy in a contextdependent fashion. It is possible that different sensitivity thresholds, the exact nature of which remain to be determined, can dictate whether autophagy or apoptosis will develop. Alternatively, the choice between apoptosis and autophagy is influenced by the fact that the two catabolic processes exhibit some degree of mutual inhibition. In some cases, a mixed phenotype of apoptosis and autophagy can be detected at the single-cell level. Although autophagy mostly allows cells to adapt to stress, massive autophagy can also kill cells. Regulation of autophagy and apoptosis by p53 and p19ARF/p14ARF Oncogenic stress can activate two different isoforms of murine p19ARF (and of the human orthologue p14ARF). One isoform corresponds to the full-length nucleolar p19ARF/p14ARF and the second one corresponds to a shorter form, smARF, which results from internal initiation of translation. Whereas smARF translocates to mitochondria, reduces the mitochondrial transmembrane potential (Dm) and stimulates autophagy and caspase-independent cell death, the nucleolar p19ARF/p14ARF activates p53 by binding and inhibiting the negative regulator of p53, MDM2. p53 induces caspase-dependent apoptotic cell death via the induction of multiple pro-apoptotic proteins, most of which function in the intrinsic pathway. Whereas the nucleolar pathway is inhibited by Z-VAD, smARF-induced cell death is blocked by knocking down beclin-1 (Becn1) or Atg5. In some cellular settings, activated p53 induces a transcriptional programme that favours the accumulation of autophagic vacuoles via the induction of the lysosomal protein DRAM (damaged-regulated autophagy modulator). p53-elicited DRAM may be essential, both for autophagic vacuolization and for apoptosis. BH3 proteins and mimetics act on the beclin-1–BCL2 interaction • Proteins that contain BCL2 homology-3 (BH3) domains or small molecules that mimic BH3 domains can bind to the BH3-receptor domain of BCL2 or BCL-XL and, hence, competitively disrupt the interaction between BCL2 or BCL-XL and beclin-1. This probably leads to the activation of the lipid kinase activity of the class III phosphatidylinositol 3-kinase VPS34 (which depends on the interaction with UVRAG (UV irradiation resistance-associated tumour suppressor gene), thereby provoking the production of phosphatidylinositol-3-phosphate (PtdIns3P). In turn, this leads to vesicle nucleation in a manner that probably involves WIPI-1a (WDrepeat protein interacting with phosphoinositides). Molecular switches between apoptosis and autophagy a | Dual function of autophagy protein-5 (ATG5) in autophagy. Full-length ATG5 can participate in the initial stages of autophagy with the help of a ubiquitin-like conjugation system. ATG12 (activated by ATG7 and then transferred to ATG10) is conjugated with ATG5 at a lysine-amino group, allowing it to bind to ATG16. The resulting ATG12–ATG5–ATG16 complex imposes curvature on the crescent phagophore and recruits activated LC3 to the elongating membrane (LC3 is one of the mammalian homologues of Atg8, which is conjugated to phosphatidylethanolamine by the action of ATG4, ATG7 and ATG3) (Box 1). Proteolysis by a calpain results in a truncated ATG5, which can translocate to mitochondria and induce MOMP. b | BCL2 homology-3 (BH3)-only proteins as inducers of apoptosis and autophagy. Depending on their specificity (for pro- or antiapoptotic and autophagy-inhibitory proteins that contain BH3 receptors), as well as their preferential subcellular localization (mitochondria or endoplasmic reticulum), BH3only proteins can preferentially activate apoptosis or autophagy. A specific class of inactivator BH3-only protein interacts with antiapoptotic and anti-autophagic multidomain proteins of the BCL2 family (red blocking arrows symbolize preferential affinities), thereby releasing activator BH3-only proteins that allosterically activate BAX and BAK (for the induction of apoptosis). Alternatively, they release beclin-1 from its inhibitory interaction with multidomain proteins of the BCL2 family, allowing it to activate the lipid kinase activity of VPS34 and, hence, to activate autophagy. Several steps in this working model still need further experimental validation. Scenarios for the interaction between apoptosis and autophagy • a | Schematically, a particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards necrosis (parts 2,3). b | Some conditions can trigger a lethal autophagic response that is responsible for cell death, for example, in naïve cells (parts 1,2) or in cells in which the apoptotic pathways have been interrupted. c | Another set of stimuli (or perhaps simply a lower dose of insults) provokes a protective autophagic response (part 1), which is required for adaptation of the cell and the avoidance of apoptosis (part 2). d | Frequently, lethal conditions trigger an autophagic response that, independently of the autophagic response, is followed by apoptosis (part 1). In this case, inhibition of apoptosis causes either cell survival (part 2) or necrosis (part 3). In this scenario, the order of events (autophagy, then apoptosis) is chronological, not hierarchical, meaning that inhibition of autophagy does not prevent apoptosis. e | Autophagy can be indispensable for sustaining the high ATP levels that are required for cells to emit signals to phagocytic cells that engulf the apoptotic bodies (part 1). Inhibition of autophagy does not affect apoptotic cell death, yet it abolishes the heterophagic removal of apoptotic material (part 2). LPC, lysophosphatidylcholine; PS, phosphatidylserine. • Eukaryotes have two major protein degradation systems within cells. One is the ubiquitin-proteasome system, which accounts for the selective degradation of most short-lived proteins. The other is the lysosomal system. Autophagy (Macroautophagy) is the primary means for the degradation of cytoplasmic constituents in the lysosome. Intracellular protein degradation systems (Upper) Macroautophagy. A small volume of cytoplasm is enclosed by the autophagic isolation membrane, which eventually results in the formation of an autophagosome. The outer membrane of the autophagosome then fuses with the lysosome where the cytoplasm-derived materials are degraded. (Lower) The ubiquitin-proteasome pathway. Polyubiquitinated proteins are degraded by the 26S proteasome. This is a selective process. • Cellular events in autophagy The cellular events during digestion of self constituents or intracellular pathogens follow three distinct stages: initiation (formation of the phagophore), elongation (growth and closure) and maturation of a double membrane autophagosome into an autolysosome. a | Autophagy sequesters and removes cellular constituents from the cytosol, including surplus or damaged organelles from the cytosol. b | Autophagy can eliminate bacteria (free in the cytosol or inside a phagosome), viruses and protozoan parasites in a manner similar to the elimination of self constituents. The cellular process of autophagy. • Conditions such as nutrient starvation, pathogen infection and other environmental stressors, can induce autophagy. Autophagy begins with the isolation of double-membrane-bound structures inside an intact cell. Previously, these structures were believed to be derived from the ribosome-free region of the rough endoplasmic reticulum82, 83, but recent studies indicate that they might originate from a pre-existing membrane structure called a phagophore84, or could be formed de novo80, 85. These membrane structures elongate and mature, and microtubule-associated protein 1 light chain 3 (LC3) is recruited to the membrane. The elongated double membranes form autophagosomes, which sequester cytoplasmic proteins and organelles such as mitochondria. The formation of the pre-autophagosomal structure can be inhibited by the phosphatidylinositol 3phosphate kinase (PI3K) inhibitor 3-methyladenine (3-MA). Sequestration requires ATP and is regulated mainly by class III PI3K46. The autophagosomes mature with acidification by the H+-ATPase71 and fuse with lysosomes to become autolysosomes (also known as the degradative autophagic vacuoles). Microtubules are important mediators of this fusion process. This process is inhibited by the H+ATPase inhibitor bafilomycin A1, or by microtubule inhibitors such as vinblastine and nocodazole86, 87. Eventually, the sequestered contents are degraded by lysosomal hydrolases for recycling. One assay for autophagic cells is to detect the presence of membrane-bound LC3, which accumulates on autophagosomes. Autophagy, meaning ‘to eat oneself’, is one of the main mechanisms for maintaining cellular homeostasis. This pathway is not directly a death pathway, rather a selfcannibalisation pathway. Mediated via the lysosomal degradation pathway, autophagy is responsible for degrading cellular proteins and is currently the only known process for degrading cellular organelles, recycling them to ensure cell survival. Research on autophagy has been on-going for over 40 years, but has been restricted by lack of knowledge about the molecular machinery behind this process. Over the last decade, huge advances have been made and genetic screens in yeast (s. cerevisiae) have led to the identification of over ~30 autophagy-related genes (ATGgenes), many of which have identified mammalian homologues (see table below) (1,2,3,4). Autophagy is essential in helping to maintain the balance between the increase and decrease in the number of a cell population. It is undoubtedly active at a basal level in most cells and contributes to the routine turnover of cytoplasmic components (1, 21). Table of identified autophagy-related genes (ATG-genes) TABLE 1 | Autophagy genes in mammals Gene Protein function ATG1, ULK1‡ Atg1 is a serine/threonine protein kinase; it may be involved in regulation and vesicle formation ATG3 Atg3 functions as an ubiquitin-conjugating-like enzyme that covalently attaches Atg/LC3 to phosphatidylethanolamine ATG4 Atg4 is a cysteine protease that cleaves the C-terminus of Atg8/LC3 to expose a glycine residue for subsequent conjugation ATG5 Atg5 is covalently attached to Atg12, and binds Atg16 as part of a tetrameric complex of unknown function ATG6, Beclin 1‡ Atg6 is a component of the class III phosphotidylinositol-3-kinase complex that is required for autophagy ATG7 Atg7 is a homologue of the ubiquitin-activating enzyme; it activates both Atg8/LC3 and Atg12 before conjugation ATG8, MAP1LC3‡ Atg8/LC3 has structural similarity to ubiquitin; it is conjugated to phosphatidylethanolamine, and is part of the autophagosome, but its function is not known ATG9 Atg9 is a transmembrane protein that may be involved in delivering membrane to the forming autophagosome ATG10 Atg10 functions as an ubiquitin-conjugating-like enzyme that covalently attaches Atg12 to Atg5 ATG12 Atg12 has some structural similarity to ubiquitin; it is conjugated to an internal lysine of Atg5 through its C-terminal glycine ATG16 Atg16 binds Atg5 and homo-oligomerizes to form a tetrameric complex *In this table we have only listed certain key genes in which the gene product has been confirmed to play a role in autophagy in higher eukaryotes. ‡Only the autophagy-related gene (ATG) name and number is listed except when that name is not used in higher eukaryotes. However, many of the autophagy genes are present as multiple isoforms, usually denoted as a, b and so on. TABLE 2 | Known small molecules that influence autophagy Compound Target Effect Reference s Rapamycin Target of rapamycin Induces autophagy 33,34 Lithium, sodium valproate, carbamezapine Enzymes that ultimately affect myo-inositol-1,4,5triphosphate levels, such as inositol monophosphatase Lowers myo-inositol-1,4,5triphosphate levels, induces autophagy 41 3-methyladenine Class III phosphotidylinositol-3kinase Inhibits autophagy 92 Wortmannin Class III phosphotidylinositol-3kinase Inhibits autophagy 93 Bafilomycin A1 Vacuolar-ATPase Inhibits autophagy 81,82 Chloroquine Lysosomal pH Inhibits autophagy 71 Hydroxychloroquine Lysosomal pH Inhibits autophagy 83 Tamoxifen Beclin 1 Increases Beclin 1, induces autophagy 70 • Figure 1: Steps of autophagy and associated genes. Shown are the three steps of autophagy accompanied by the genes associated with each step. Genes listed in green have already been assessed by Ray Choi, genes listed in red will be the target of assessment in this paper and genes listed in black remain to be assessed in the future. • The term ‘Autophagy’ covers three processes; microautophagy, macroautophagy and chaperonemediated autophagy (1): • Microautophagy is the transfer of cytosolic components into the lysosome by direct invagination of the lysosomal membrane and subsequent budding of vesicles into the lysosomal lumen (17). • Macroautophagy involves formation of a double-membrane structure called the autophagosome which sequesters cytosolic material and delivers it to the lysosome for degradation (18). Although this degradation can be selective (i.e. specific removal of damaged mitochondria sparing normal functioning ones) (19), degradation of soluble cytosolic proteins is non-selective. • Chaperone-mediated autophagy (CMA) is characterised by its selectivity regarding the specific substrates (cytosolic proteins) degraded (20). Autophagy - the lysosomal system • • • • • In general terms, 'autophagy' refers to any intracellular process that involves the degradation of cytosolic components by the lysosome. There are at least three distinct autophagic pathways: Macroautophagy, Microautophagy and Chaperone-mediated autophagy. The terms autophagy and macroautophagy are used interchangeably. Autophagy is involved in physiological and pathological processes Autophagy is a lysosomal degradation pathway that is essential for survival, differentiation, development, and homeostasis. Under certain circumstances, it is considered as a non-apoptotic cell death pathway. Autophagy is involved in diverse pathologies, including infections, cancer, neurodegeneration and aging as well as in heart, liver and kidney diseases. The morphological hallmark of autophagy Macroautophagy is a multistep process by which portions of cytoplasm, damaged proteins and/or organelles are sequestered in a double or multimembrane structure (autophagosome). Upon fusion of the autophagosome with the lysosome (autophagolysosome), sequestered materials are digested by lysosomal hydrolases. The recycling of these intracellular constituents serves as an alternative energy source during periods of metabolic stress to maintain homeostasis and viability. • • Steps in macroautophagy and chaperone-mediated autophagy (CMA). Macroautophagy: 1.) Nucleation. An unidentified membrane source delivers lipid bi-layers for the formation of the phagophore. In yeast this early structure is termed pre-autophagosomal structure (PAS), its identity in mammalian cells is uncertain. A class III PI3K complex consisting of at least BECN1, PIK3C3, PIK3R4, UVRAG, and AMBRA1 is required for PAS formation and MAP1LC3 is anchored to the membrane via a phosphoethanolamine (PE) anchor (LC3-II). 2.) Expansion. The PAS or a comparable structure in mammals sequesters cytosolic cargo (either specifically via SQSTM1 [p62] or nonspecifically) by invagination, forming a double-membranous vesicle. This stage is also called "isolation membrane". More membrane and LC3-II is being recruited to the developing vacuole. 3.) Maturation. The completed autophagosome undergoes multiple maturation steps and fusion events with multi-vesicular bodies (MVB) or endosomes. The exact nature and sequence of this maturation, and whether these steps are always required is currently unknown. The autophagosomal lumen becomes more acidified during this maturation. 4.) Docking and fusion. During docking and fusion the inner membrane compartment together with its content gets released into the lysosome/autolysosome and is being degraded by lysosomal hydrolases. The components of the outer membrane are available for re-usage. Chaperone-mediated autophagy: 5.) Recognition and binding. The HSC70 chaperone complex (consisting of HSC70, HSP90 and maybe other proteins) recognizes unfolded proteins with the KFERQ sequence and moves them to the lysosome. 6.) Translocation. LAMP2A and a lysosomal form of HSC70 (l-HSC70) translocate the substrate protein across the lysosomal membrane into the lumen for degradation. The autophagy delivered substrates get degraded inside the lysosomes and their macromolecular components are made available to the cell's metabolism via permeases that allow their transport back into the cytosol. Jaeger and Wyss-Coray Molecular Neurodegeneration 2009 4:16 doi:10.1186/1750-1326-4-16 • • • Macroautophagy Macroautophagy is a nearly universal process that eukaryotic cells employ to reutilize the constituents of cytoplasm and organelles. We have used the genetics of Dictyostelium to isolate mutants in genes that are essential for macroautophagy. These mutants have taught us how to design a screen and a selection for other genes that regulate macroautophagy and also provide material for studying autophagosome formation. A collection of mutants has been used to show that macroautophagy, previously thought to be essential for replication of the intracellular pathogen Legionella pneumophila, is not required for growth of this parasite. We now are asking how autophagy is linked to development and whether there are additional genes that are essential for autophagy. Macroautophagy: Macroautophagy as a cell biological and structural phenomenon has been known for many years to manage the bulk degradation of cytoplasm and organelles. One of its interesting features is that it promotes the transfer of material from one topologically distinct compartment to another - from the cytosol to the vacuole in baker yeast or to lysosomes in other eukaryotic cells. An initiating structure called the Pre-autophagosomal Structure or PAS has been defined, but its origins, once thought to be from the endoplasmic reticulum, are not clear. The favorable vacuolar structure of Saccharomyces cerevisiae has been married to the genetics of that organism to understand the biochemical reactions involved in the production of autophagosomes (for reviews see (Abeliovich and Klionsky, 2001; Klionsky and Emr, 2000; Thumm, 2000). S. cerevisiae contains a single large central vacuole, which serves as lysosome, among other functions. When autophagy is induced, double-membrane autophagosomes are produced, and these fuse with the vacuole and release single membrane autophagic bodies that are degraded by resident hydrolases and proteases. When vacuolar proteases are inhibited by PMSF, one observes an accumulation of small spheres (autophagic bodies) within a larger vacuole. Using several screening methods to create mutants in which an accumulation of the small autophagic bodies is not observed, several laboratories, notably those of Ohsumi, Thumm, and Klionsky, have isolated autophagy mutants (called atg). The affected genes have been used to define a series of phosphorylation and ubiquitination-like reactions that are necessary for cytoplasm to vesicle transport (CVT) and macroautophagy. CVT encapsulates specific targets during growth, while autophagy envelopes bulk cytoplasm and organelles during starvation. Many of the molecules that mediate autophagy reside in the Pre-autophagosomal structure (PAS, also called the perivacuolar compartment (PVC)), which in budding yeast lies next to the vacuole (Suzuki et al., 2001) (Kim et al., 2002) (Noda et al., 2002). Gradient and immuno-purification studies suggest that the PAS does not contain markers from other compartments of the cell (Kim et al., 2002). The creation of an autophagosome is thought to occur by extension of an enveloping membrane, called an isolation membrane, which then closes around cytoplasmic constituents, as shown in Figure 1 (Kim et al., 2002). A similar situation may occur in mammalian cells (Mizushima et al., 2001). Autophagosomes then fuse with late endosomes or lysosomes to create autophagolysosomes and provide constituents and energy for the cell. Although there has been progress in understanding this process, especially in determining the genes involved and mapping known gene products onto physical structures, a number of questions remain. What is the origin of the PAS and of what is it composed, beyond the known molecules? Does it assemble de novo or does it derive from pre-existing membranes? How does the PAS convert into an isolation membrane and how does that membrane seal around its cargo? What is the source of the lipid that forms the membrane? How is size determined? Is there selectivity of cargo? What are the molecules that mediate fusion with the endosomal system? Schematic overview of macroautophagy. Potential therapeutic applications of autophagy David C. Rubinsztein, Jason E. Gestwicki, Leon O. Murphy & Daniel J. Klionsky Nature Reviews Drug Discovery 6, 304-312 (April 2007) • A membrane of unknown origin forms the initial phagophore or isolation membrane. The phagophore expands, sequestering cytoplasm and on completion forms a doublemembrane autophagosome. The autophagosome fuses with a lysosome, containing acid hydrolases (AH), which can now gain access to the inner vesicle, termed an autophagic body. The fused compartment where the autophagic body and its contents are degraded is called an autophagolysosome or autolysosome. Following breakdown, the resulting macromolecules are released back into the cytosol through permeases for reuse in metabolic processes (during starvation). Alternatively, the cargo might be inactivated or killed (when macroautophagy acts as part of the immune response to eliminate microbial pathogens), or the removal of cytoplasm might result in cell death. See main text for details. It is a conceptually simple, intellectually appealing, and experimentally testable hypothesis. Since joining the faculty of the Pharmacology Department of RWJMS in 2003, my laboratory has conducted extensive research around this basic theme. We are working in four interrelated research areas and have been making significant progress. First, we are seeking experimental proof that a defect in autophagy compromises mitochondrial functions, a leading cause of aging. Second, we are determining the direct cause of cancer development as a result of reduced autophagy. Third, we are trying to understand the regulation of autophagy. Fourth, we are exploring the potential of targeting autophagy as a therapeutic adjuvant to cancer therapy. In addition to the Arnold Levine Laboratory, where I was trained as a postdoc, we have made several UMDNJ laboratories our scientific allies along the way, including the labs of Eileen White, William Hait, and Leroy Liu. These various collaborations have helped move the science forward far more quickly. Three hundred years ago, most people died before the age of 50. Few suffered from cancer, dementia, or type II diabetes. Since the Industrial Revolution, the average lifespan of human beings has increased dramatically. Alas, so has the suffering associated with these aging-related diseases. If autophagy is indeed involved in aging control and preventing those agingrelated diseases, understanding autophagy might reveal the secret that prolongs the human lifespan without increasing suffering. Are we looking for the “Holy Grail”? Yes, that is our dream. It turned out that this strain of mouse, which contains a deletion in a gene called beclin1, has a much higher chance of getting cancer when it is old. In other words, this result demonstrates that the normal function of the beclin1 gene is to prevent tumor development in old age. Interestingly, the human version of the beclin1 gene is also deleted in sporadic human cancers, tumors affecting only older people. Approximately 50% of sporadic breast cancers, 75% of sporadic ovarian cancers, and 40% of sporadic prostate cancers contain a deletion of the human version of the beclin1 gene. What is even more interesting is that the gene is not related to any known processes that cause cancer. Instead, this gene participates in autophagy, an evolutionarily conserved process that is involved in recycling cytoplasmic components. As shown in Figure 1, autophagy starts with the emergence of a crescent, double membrane structure, which grows and engulfs a portion of cytoplasm, including mitochondria, ribosomes, and proteins. The structure then matures to a closed, double membrane vesicle with a diameter between 0.3 and 0.9 micrometers, which is called an autophagosome or autophagic vacuole. The autophagosome then travels toward the lysosome, a garbage depot and recycling center in the cell. The outer membrane of the autophagosome is then fused with the lysosome membrane, leading to the release of its cargo, which is wrapped by the inner membrane into the lysosome. The cargos are then degraded by the various enzymes in the lysosome. Basically, it is believed that one function of autophagy is to serve as sanitation workers busily collecting and recycling garbage in the cells. Strikingly, the same autophagy process has been demonstrated to be important in regulating the life-spans of the flatworm C. elegans, a common and convenient animal model used in the biological laboratory. The laboratory of Beth Levine, MD, at Southwest Medical Center, first cloned the beclin1 gene and independently demonstrated that a mouse strain lacking the beclin1 gene has higher cancer rates, and Leroy Liu, PhD, chair of the Pharmacology Department at UMDNJ-Robert Wood Johnson Medical School (RWJMS), demonstrated that inactivation of autophagy genes shortens the worms’ lives. In the meantime, other scientists, including Zhenyu Yue, PhD, who is now an assistant professor at Mount Sinai School of Medicine in New York and works on the neurobiological aspects of autophagy, found that some aging-related neurodegenerative diseases caused by the abnormal accumulation of protein aggregates in neurons, such as Huntington’s disease, are associated with autophagy reduction. The pharmacological induction of autophagy can help clear the disease-causing aggregates in cells and alleviate the symptoms of these diseases. An exciting theme then emerged: Autophagy is a critical garbage collecting and recycling process in healthy cells. As we get older, the process slows down or becomes less discriminating. Consequently, haphazard agents are accumulated in cells, which damage various parts of cells and tissues, leading to some aging-related diseases. For example, failure to clear protein aggregates in neurons of the central nervous system causes dementia; failure to clear ROS (reactive oxygen species)-producing mitochondria leads to nuclear DNA mutations and cancer. Collectively, these pathological conditions contribute to, or even define, the process of aging. Figure 1 Specific and nonspecific autophagy From the following article: Autophagy: from phenomenology to molecular understanding in less than a decade Daniel J. Klionsky Nature Reviews Molecular Cell Biology 8, 931-937 (November 2007) • • During nonspecific autophagy (see figure part a), sequestration begins with the formation of a phagophore that expands into a double-membrane autophagosome while surrounding a portion of the cytoplasm. The autophagosome may fuse with an endosome (the product of endocytosis), which is a form of heterophagy (in a heterophagic process, the cell internalizes and degrades material that originates outside of the cell, in contrast to autophagy, in which the cell consumes part of itself). The product of endosome–autophagosome fusion is known as an amphisome. The completed autophagosome or amphisome fuses with a lysosome, which supplies acid hydrolases. The enzymes in the resulting compartment, an autolysosome, break down the inner membrane from the autophagosome and degrade the cargo. The resulting macromolecules are released through permeases and recycled in the cytosol. The yeast cytoplasm-to-vacuole targeting (Cvt) pathway is one example of specific autophagy (see figure part b), and the only example of a biosynthetic autophagy-related pathway. The overall morphology is identical to nonspecific autophagy; however, the sequestering Cvt vesicles are smaller than autophagosomes and they appear to exclude bulk cytoplasm. The phagophore assembly site (PAS) either becomes the sequestering vesicle or generates it. The precursor form of aminopeptidase I (prApe1) forms oligomers in the cytosol, and is targeted through the action of a receptor, Atg19, and the adaptor or scaffold protein Atg11 to allow selective cargo recognition and packaging. The completed vesicle fuses with the vacuole, the yeast analogue of the mammalian lysosome. The larger size of the vacuole allows the release of the inner single-membrane compartment of the Cvt vesicle, which is now termed a Cvt body; during nonspecific autophagy in yeast, this structure is termed an autophagic body. The Cvt body is lysed and prApe1 is matured by proteolytic removal of a propeptide to generate the active, resident hydrolase mApe1. Regulation of autophagy in mammalian cells From the following article: Autophagy: from phenomenology to molecular understanding in less than a decade Daniel J. Klionsky Nature Reviews Molecular Cell Biology 8, 931-937 (November 2007) • In the figure, the green circles represent components that stimulate autophagy, whereas the purple boxes correspond to inhibitory factors. 3-methyladenine (3-MA) and wortmannin (Wm) also inhibit class I phosphatidylinositol 3-kinases (PI3K), but the overall effect of these compounds is a block in autophagy (because they inhibit the downstream class III enzyme that produces phosphatidylinositol-3phosphate (PtdIns(3)P), which is needed for autophagy). The regulation of autophagy is complex and far from understood. Historically, TOR (target of rapamycin) has been considered to be the central regulator of autophagy, because TOR inhibition with rapamycin (Rap) induces autophagy. However, it is now clear that there are also TOR-independent types of regulation. For example, beclin-1 and Atg4 might be regulated by the c-Jun N-terminal kinase (JNK) and reactive oxygen species (ROS), respectively. Additional information is available in recent reviews (for example, see Refs 74, 75). • Perspectives • Nature Reviews Molecular Cell Biology 8, 931-937 (November 2007) Autophagy: from phenomenology to molecular understanding in less than a decade • Daniel J. Klionsky • Abstract • In 2000, it was suggested to me that “Autophagy will be the wave of the future; it will become the new apoptosis.” Few people would have agreed at the time, but this statement turned out to be prophetic, and this process of 'self-eating' rapidly exploded as a research field, as scientists discovered connections to cancer, neurodegeneration and even lifespan extension. Amazingly, the molecular breakthroughs in autophagy have taken place during only the past decade. • • • Autophagy, the process by which proteins, organelles and invading pathogens are sequestered in autophagosomal vesicles and delivered to the lysosome for degradation, provides a primary route for turnover of stable, defective and unwanted cellular components. Defects in this cellular homeostasis system are linked with numerous human diseases. This process involves a series of dynamic membrane-rearrangement reactions mediated by a core set of autophagy (Atg) proteins. Although conserved protein kinase, lipid kinase and ubiquitin-like protein conjugation subnetworks controlling autophagosome formation and cargo recruitment have been defined, several gaps remain in our understanding of autophagy. For instance, it has been unclear how the distinct protein complexes involved in autophagy talk to each other or to other cellular machinery involved in membrane formation and membrane trafficking. And how do key signals that trigger autophagy talk to the Atg proteins? Autophagy networks. (Top) Overview of macroautophagy. (Bottom) Major signal transduction systems in autophagy. Recently, we have performed a systematic proteomic analysis of the human autophagy system with the long-term goal of providing molecular insight into some of the unanswered questions in the field. This effort has provided a glimpse into the global architecture of the autophagy interaction network (AIN). Briefly, 65 autophagy-related proteins have been analyzed proteomically by IP-coupled LC-MS/MS, revealing a dense interaction network with extensive cross-talk among functional signaling modules. The current AIN contains many new components linked to vesicle trafficking, ATG8 conjugation, protein phosphorylation, protein ubiquitination, and lipid modification, and together with genetic (RNAi) and localization studies performed in parallel, provides a framework for understanding the molecular architecture of the pathway and how individual sub-networks communicate with each other. Complementary, we carried out detailed analyses of ATG8 interaction proteins to delineate regulators and cargo of selective autophagy. Together, our work provides a resource for detailed mechanistic and further systematic analysis of this critical protein homeostasis pathway. • Three predominant cellular functions can be assigned to autophagy: • Autophagy is a response to nutrient starvation. Decreased levels of amino acids can induce the autophagic response in numerous cell types and situations e.g. the neonatal period, when the supply of nutrients via the milk has not yet replaced the nutrients via the placenta (6,7) • Autophagy is a housekeeping process whereby long-lived proteins and organelles are recycled e.g. Mitochondria (22) • Autophagy has tissue-specific roles e.g. during erythrocyte development, following nucleus expulsion, autophagy is required to degrade the remaining organelles. Degradation of the autophagic vesicle results in the functional biconcave shape (23, 24). • The Four Stages of Autophagy Induction: Following external/internal stimuli (e.g. nutrient depletion or ischemia), mTOR is inhibited, leading to induction of autophagy. • Autophagosome formation: Cytosolic proteins and organelles are sequestered by a double membrane vesicle, the origin of which is uncertain, but may arise from the endoplasmic reticulum. Formation of this vesicle is co-ordinated by complexes of Atg proteins, which encode for the “start codons,” which encode for the amino acid methionine. • Docking and fusion with the lysosome. • Breakdown of the autophagic vesicle. The molecular mechanism behind the fusion with the lysosome and subsequent breakdown of the autophagic vesicle are poorly understood. Schematic diagram of the steps of autophagy. Autophagy begins with the formation of the phagophore or isolation membrane (vesicle nucleation step). The concerted action of the autophagy core machinery proteins at the phagophore assembly site (PAS) is thought to lead to the expansion of the phagophore into an autophagosome (vesicle elongation). The autophagosome can engulf bulk cytoplasm nonspecifically, including entire organelles, or target cargos specifically. When the outer membrane of the autophagosome fuses with an endosome (forming an amphisome before fusing with the lysosome) or directly with a lysosome (docking and fusion steps), it forms an autophagolysosome. Finally, the sequestered material is degraded inside the autophagolyosome (vesicle breakdown and degradation) and recycled. Human autophagy Signalling regulation of mammalian autophagy. • In the figure, the blue components represent the factors that stimulate autophagy, whereas the red ones correspond to inhibitory factors. Autophagy is regulated by a complex signalling network of various stimulatory (blue arrows) and inhibitory (red bars) inputs. TOR plays a central role in autophagy by integrating the class I PtdIns3K signalling and amino acid-dependent signalling pathways. Activation of insulin receptors stimulates the class I PtdIns3K complex and small GTPase Ras, leading to activation of the PtdIns3K–PKB–TOR pathway. PKB phosphorylates and inhibits the tuberous sclerosis complex 1/2 (TSC1–TSC2), leading to the stabilization of Rheb GTPase, which in turn activates TOR, causing inhibition of autophagy. Amino acids inhibit the Raf-1–MEK1/2–ERK1/2 signalling cascade, leading to inhibition of autophagy. Energy depletion causes the AMP-activated protein kinase (AMPK) to be phosphorylated and activated by LKB1. AMPK phosphorylates and activates TSC1–TSC2, leading to inactivation of TOR and autophagy induction. p70S6K kinase is a substrate of TOR that may negatively feed back on TOR activity, ensuring basal levels of autophagy that are important for homeostasis. JNK1 and DAPK phosphorylate and disrupt the association of antiapoptotic proteins, Bcl-2 and Bcl-XL, with Beclin 1, leading to the activation of the Beclin 1-associated class III PtdIns3K complex and stimulation of autophagy. Beclin 1 is shown bound to the phagophore membrane. Schematic depiction of autophagy. From Eaten alive: a history of macroautophagy Zhifen Yang1 Daniel J. Klionsky1 Journal name: Nature Cell Biology Volume: 12, Pages: 814–822 (2010) • (a) In yeast, both autophagy and the Cvt pathway engulf cargoes within distinct double-membrane vesicles, which are thought to originate from the phagophore assembly site (PAS). The PAS is defined as the initial site for autophagy-related (Atg) protein recruitment. The Cvt pathway is one example of selective autophagy, and the only example of a biosynthetic autophagy-related pathway. The Cvt vesicle (140–160 nm in diameter) appears to closely enwrap the specific cargo — the Cvt complex (consisting of the precursor form of aminopeptidase I — prApe1 — and the Atg19 receptor), and exclude bulk cytoplasm. The autophagosome (300–900 nm in diameter) engulfs cytoplasm, including organelles, and also the Cvt complex. The completed vesicles then fuse with the vacuole, the yeast analogue of the mammalian lysosome, and release the inner single-membrane vesicle (autophagic or Cvt body) into the lumen. Subsequent breakdown of the inner vesicles allows the maturation of prApe1 and the degradation of cytoplasm, and hence the recycling of the resulting macromolecules through vacuolar permeases. (b) Mammalian autophagy is initiated by the formation of the phagophore, followed by a series of steps, including the elongation and expansion of the phagophore, closure and completion of a double-membrane autophagosome (which surrounds a portion of the cytoplasm), autophagosome maturation through docking and fusion with an endosome (the product of fusion is known as an amphisome) and/or lysosome (the product of fusion is known as an autolysosome), breakdown and degradation of the autophagosome inner membrane and cargo through acid hydrolases inside the autolysosome, and recycling of the resulting macromolecules through permeases. So far, there is no evidence for a PAS that exists in mammalian cells, and so the mammalian phagophore could be equivalent to the yeast PAS, or derived from the PAS. The core molecular machinery is also depicted, such as the ULK1 and ULK2 complexes that are required for autophagy induction, class III PtdIns3K complexes that are involved in autophagosome formation, mammalian Atg9 (mAtg9) that potentially contributes to the delivery of membrane to the forming autophagosome and two conjugation systems, the LC3-II and Atg12–Atg5–Atg16L complex, which are proposed to function during elongation and expansion of the phagophore membrane. Schematic diagram of the presumed role f or C. elegans autophagy proteins involved in the formation of an autophagosome. A) Regulation of induction: In yeast, the Tor kinase and its effectors regulate the induction of autophagy. UNC-51 is the C. elegans Atg1 ortholog, however, it is not clear if there are similar regulatory proteins to Atg17 or Atg13. B) Vesicle nucleation requires a lipid kinase complex, which includes the class III phosphatidylinositol 3-kinase (PI3K), Vps34 (in C. elegans LET-512). In yeast, Vps34 activation depends on its binding partners, Atg6, Atg14, and Vps15 (Kihara et al., 2001). In C. elegans, the ATG6 ortholog is bec-1, however, orthologs to ATG14 and VPS15 appear to be missing in the C. elegans genome. The interaction between the antiapoptotic protein CED-9 and BEC-1 is conserved in C. elegans (TakácsVellai et al., 2005). A similar interaction between the mammalian proteins Bcl-2 and Beclin 1, inhibits autophagy (Maiuri et al., 2007a; Maiuri et al., 2007b; Pattingre et al., 2005). C) Two novel ubiquitin-like conjugation pathways: the Atg12 conjugation system (Atg5, Atg12, and Atg16), and the Atg8 lipidation system (Atg8, Atg3, and Atg7) mediate vesicle expansion, and vesicle completion. Orthologs to all the members of these two complexes have been found in C. elegans, where they are named ATG-3, ATG-4, ATG-5, ATG-7, ATG-16, LGG-1/Atg8 and LGG-3/Atg12. In yeast, Atg8 undergoes two posttranslational processing events resulting in conjugation to phosphatidylethanolamine (PE) and recruitment to the PAS membrane. ATG-7 is an E1 ubiquitin activating enzyme required for the activation of of LGG-1. LGG-3-ATG-5 oligomerize with ATG-16 to allow for the formation of the multimeric complex. ATG-3 and ATG-10 are E2-like ubiquitin conjugating enzymes and ATG-4 is a cysteine protease. As for its yeast ortholog, LGG1 appears to remain in the completed autophagosome and thus is an excellent marker for early and late autophagosomal structures. D) The retrieval of the integral membrane protein ATG-9 from the phagophore assembly site (PAS) involves ATG-2 and ATG18, two interacting peripheral proteins • • • • • • • The four stages of Autophagy 1) Induction: Following external/internal stimuli (e.g. nutrient depletion or ischemia) mTOR is inhibited, leading to induction of Autophagy. Key genes in yeast are Atg1 and Atg13, for which the mammalian homologues are yet to be identified. 2) Autophagosome formation: Cytosolic proteins and organelles are sequestered by a double membrane vesicle, the origin of which is uncertain, but may arise from the endoplasmic reticulum. Formation of this vesicle is co-ordinated by complexes of Atg proteins, in particular Atg5 and Atg12, that are conjugated enabling the recruitment of LC3 (Atg8). Beclin-1 forms a complex with Atg14. 3) Docking and fusion with the lysosome 4) Breakdown of the autophagic vesicle. The molecular mechanism behind the fusion with the lysosome and subsequent breakdown of the autophagic vesicle are poorly understood, although Lamp-2 is thought to play a key role. Induction – Autophagy can be induced by both internal and external stimuli. Autophagy is inhibited under nutrient-rich conditions and therefore simulated by starvation (25, 26). A key regulator or gatekeeper of autophagic induction is Tor/mTOR (14, 27, 28). In nutrient-rich conditions, mTOR has an inhibitory effect on autophagy, under starvation conditions, mTOR is inactivated – leading to the inhibition of autophagy being released. Tor inactivation leads to downstream dephosphorylation events resulting in transcriptional activation of autophagy genes (1, 28). Autophagosome formation – Following induction, a double membrane vesicle forms in the cytosol, sequestering those cytoplasmic components for autophagic degradation. The mechanism for the formation of this membranous vesicle is not well defined although it is known that most of the ATG genes identified to date participate in the formation of this vesicle (29). It has been postulated that the endoplasmic reticulum is the origin of this membrane (30, 31) • • Autophagosome fusion - During this phase, the autophagosome fuses with the lysosome, as a result the contents of the autophagosome are released into the lysosome for degradation by lysosomal proteases (1, 32). Autophagosome breakdown – Following fusion of these two vesicle bodies, the autophagosome membrane is the broken down by the lysosomal proteases (1, 31). Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ and Rubinsztein DC. (2005) Lithium induces autophagy by inhibiting inositol monophosphatase. Journal of Cell Biology 170(7):1101-1111. Access article Supplementary information Abstract: Macroautophagy is a key pathway for the clearance of aggregate-prone cytosolic proteins. Currently, the only suitable pharmacologic strategy for up-regulating autophagy in mammalian cells is to use rapamycin, which inhibits the mammalian target of rapamycin (mTOR), a negative regulator of autophagy. Here we describe a novel mTOR-independent pathway that regulates autophagy. We show that lithium induces autophagy, and thereby, enhances the clearance of autophagy substrates, like mutant huntingtin and alpha-synucleins. This effect is not mediated by glycogen synthase kinase 3beta inhibition. The autophagy-enhancing properties of lithium were mediated by inhibition of inositol monophosphatase and led to free inositol depletion. This, in turn, decreased myo-inositol-1,4,5-triphosphate (IP3) levels. Our data suggest that the autophagy effect is mediated at the level of (or downstream of) lowered IP3, because it was abrogated by pharmacologic treatments that increased IP3. This novel pharmacologic strategy for autophagy induction is independent of mTOR, and may help treatment of neurodegenerative diseases, like Huntington‘s disease, where the toxic protein is an autophagy substrate. Feature: • • • • • Sci. STKE, 4 October 2005 Vol. 2005, Issue 304, p. tw349 NEURODEGENERATIVE DISEASE Lithium Decreases IP3 to Promote Autophagy Lithium is widely known for its application in the treatment of bipolar and unipolar mood disorders. Sarkar et al. provide evidence that lithium may also be effective in the treatment of neurodegenerative disorders arising from toxicity of the accumulation of aggregate-prone proteins, such as mutant huntingtin and mutant synuclein, which are implicated in Huntington's disease and certain forms of Parkinson's disease. Stimulation of autophagy may be a mechanism to prevent the accumulation of these aggegrate-prone proteins, and Sarkar et al. show that lithium promotes autophagy, detected as an increase in the formation of autophagic vesicles and a decrease in the abundance of known autophagic substrates. Furthermore, lithium decreased the accumulation of mutant -synuclein or huntingtin aggregates and the toxicity (cell death) of these proteins in inducible PC12 cell lines or COS-7 cell lines, an effect that was blocked by the autophagic inhibitor 3methyladenine (3-MA). Lithium is a nonspecific inhibitor of several enzymes. The effect on autophagy appeared to be mediated by the inhibition of inositol monophosphatase (IMPase), because autophagy and clearance of mutant huntingtin and -synuclein was also stimulated by inhibition of IMPase with a specific pharmacologic agent (L-690,330), whereas selective inhibition of glycogen synthase kinase-3ß (GSK-3ß, another lithium target) did not promote clearance of the aggregate-prone proteins. Both L-690,330 and lithium decreased inositol1,4,5-trisphosphate (IP3) concentrations. That lithium exerted its effects on autophagy through decreased IP3 concentrations was validated by experiments in which IP3 concentrations were increased before lithium treatment by addition of myo-inositol or prolyl endopeptidase inhibitor 2 (PEI). Increased IP3 concentration promoted the accumulation of mutant huntingtin and cell death and blocked the effects of lithium to reduce these phenomena. Autophagy is also stimulated by inhibition of mTOR (mammalian target of rapamycin); however, lithium appeared to act independently from the pathway regulated by mTOR, because inhibition of mTOR with rapamycin combined with lithium had an additive effect on clearance of the aggregate-prone proteins and cell survival. The ability of lithium to stimulate autophagy and clearance of aggregate-prone proteins was also observed for other mood-stabilizing drugs that promote inositol depletion. Thus, a pathway involving IP3 and inositol appears to be one mechanism regulating autophagy that may be exploited for therapeutic benefit in certain neurodegenerative diseases. S. Sarkar, A. Floto, Z. Berger, S. Imarisio, A. Cordenier, M. Pasco, L. J. Cook, D. C. Rubinsztein, Lithium induces autophage by inhibiting inositol monophosphate. J. Cell Biol. 170, 1101-1111 (2005). [Abstract] [Full Text] Citation: Lithium Decreases IP3 to Promote Autophagy. Sci. STKE 2005, tw349 (2005). Regulation of membrane traffic by phosphoinositide 3-kinases • Regulation of autophagy in Drosophila by PI 3-kinases. Autophagy is known to be regulated by both the class I and the class III PI 3-kinases. The class III PI 3-kinase is required for autophagy through producing PtdIns(3)P via the autophagy-specific complex containing Beclin-1, Vps15 and hVps34. (In yeast, an additional accessory protein, Vps14, has been shown to be part of the complex.) By contrast, the class I PI 3-kinase inhibits autophagy by activating the Akt/PKB pathway. The insect hormone ecdysone acts through nuclear receptors and triggers programmed autophagy in the Drosophila fat body by downregulating the class I PI 3-kinase pathway through an unknown mechanism. The overexpression of the PI 3phosphatase PTEN mimics this effect. • Sarkar S, Davies JE, Huang Z, Tunnacliffe A and Rubinsztein DC. (2007) Trehalose, a novel mTOR-independent autophagy inducer, accelerates clearance of mutant huntingtin and alpha-synuclein. Journal of Biological Chemistry 282(8):5641-5652. Access article Supplementary information Abstract: Trehalose, a disaccharide present in many non-mammalian species, protects cells against various environmental stresses. Whereas some of the protective effects may be explained by its chemical chaperone properties, its actions are largely unknown. Here we report a novel function of trehalose as an mTORindependent autophagy activator. Trehalose-induced autophagy enhanced the clearance of autophagy substrates like mutant huntingtin and the A30P and A53T mutants of alpha-synuclein, associated with Huntington disease (HD) and Parkinson disease (PD), respectively. Furthermore, trehalose and mTOR inhibition by rapamycin together exerted an additive effect on the clearance of these aggregate-prone proteins because of increased autophagic activity. By inducing autophagy, we showed that trehalose also protects cells against subsequent pro-apoptotic insults via the mitochondrial pathway. The dual protective properties of trehalose (as an inducer of autophagy and chemical chaperone) and the combinatorial strategy with rapamycin may be relevant to the treatment of HD and related diseases, where the mutant proteins are autophagy substrates. • Model of autophagosome formation in mammalian cells The Atg12-Atg5 conjugate and Apg16L localize to the isolation membrane throughout its elongation process. LC3 (Atg8 homolog) is recruited to the membrane in the Apg5-dependent manner. Apg12-Apg5 and Apg16L dissociate from the membrane upon completion of autophagosome formation, while LC3(-II) remains on the autophagosome membrane. LC3 dissociates from the autolysosomal membrane. • We generated Atg5-/- mouse embryonic stem (ES) cells and demonstrated that mammalian Atg5 is also required for autophagy. Atg5-/- ES cells can grow normally but bulk protein degradation was greatly reduced in these cells, suggesting that autophagy is actually a major degradation process. Atg5-/-EScells exhibit a block in the autophagic pathway Cells were cultured in Hanks' solution for 2 hours to induce autophagy. Bar, 1 um. (by Dr. Akitsugu Yamamoto at Nagahama Institute of Bio-Science and Technology) • In the mammalian Apg12 system, Apg12 is attached to Lys130 of Apg5. When the Atg5K130R mutant, in which Lys130 is replaced with Apg, is expressed in Atg5-/ES cells, Apg5 is no longer conjugated with Apg12. Even in such cells, Apg5K130R is able to localize to the small autophagosome precursors with Apg16L, suggesting that the covalent modification of Apg5 with Apg12 is not required for membrane targeting of Apg5 and Apg16L. However, the membrane does not elongate to form cup-shaped isolation membrane and autophagosomes. Thus, the Apg12-Apg5Apg16L complex is essential for elongation of the isolation membranes, not for the generation of the precursor structures. Atg12 conjugation is not required for membrane targeting of Atg5, but it is essential for maturation of the isolation membrane into autophagosome and recruitment of LC3 to the membrane. • • • Development of a transgenic mouse model with a fluorescent autophagosome marker Although the possible involvement of autophagy in homeostasis, development, cell death and pathogenesis has been repeatedly pointed out, systematic in vivo analysis has not been performed in mammals, mainly due to a limitation of monitoring methods. To understand where and when autophagy occurs in vivo, we have generated transgenic mice systemically expressing GFP fused to LC3, which serves as a marker protein for autophagosomes. Fluorescence microscopic analyses revealed that autophagy is differently induced by nutrient starvation in most tissues. This transgenic mouse model is a useful tool to study mammalian autophagy. • In vivo analysis of autophagy using GFP-LC3 transgenic mice (Upper) Gastrocnemius muscle samples were prepared from GFP-LC3 transgenic mice before (left) or after 24-h starvation (right). Small dots represent autophagosomes. (Lower) Embryonic fibroblasts from GFP-LC3 mice were cultured in Hanks' solution for 2 hours. Bar, 10 um. • • • Molecular Mechanism of Autophagy In the yeast, Saccharomyces cerevisiae, at least 16 of ATG genes have been identified to be required for autophagosome formation. The most surprising findings are discoveries of the two novel ubiquitylation-like conjugation systems: one mediates conjugation of Atg12 to Atg5, and the other mediates a covalent linkage between Atg8 (LC3 in mammals) and phosphatidylethanolamine (PE) (Suppl. Fig. 1)B We have dissected the autophagic process in mammalian cells using Atg homologs. We proposed a model in which the cup-shaped isolation membrane is developed from a small crescent-shaped compartment. We have also examined the localization and function of mammalian Atg proteins during this process. (A) Punctate signals of GFP-Apg5 increase under starvation conditions in ES cells. (B) Starved ES cells co-expressing CFP-Atg5 and YFP-LC3. Molecular mechanism of autophagy • Autophagy has been recently implicated in various human pathological and physiological conditions, such as neurodegeneration, immunity, cancer, development, myopathies, heart diseases, liver diseases and longevity. Basal autophagy is essential for removing misfolded proteins and damaged organelles, and therefore, plays a vital role in maintaining cellular homeostasis in all tissues. • • • • • • • Autophagy and Disease The autophagic response has been described in various pathophysiological situations, including neurobiology, cancer and more recently cardiovascular disease, however its role has yet to be teased out; the crux of the problem being, is the response a cell-protective one, or a mechanism of cell death? 3.1 Neurodegenerative diseases Studies have shown that autophagy plays a role in the removal of misfolded proteins which accumulate in the cell, such as the polyglutamine aggregates formed in Huntington’s chorea (33, 34). It could be the case though, that a disproportionate autophagic response could contribute to the pathology seen in such disorders. 3.2 Cancer A number of tumour-suppressor proteins control autophagy (e.g. Beclin-1 and PTEN), so it would seem reasonable to assume that a decrease in autophagy would lead to tumour progression (28, 35, 36). Indeed experimental evidence backs up this hypothesis - over expression of Beclin-1 induces autophagy in cultured breast cancer cells (9) and stimulation of autophagy was observed in cancer cells following anti-cancer treatments (37). Is this up-regulation of autophagy really a cell-protective mechanism, or in fact an alternative cell-death mechanism given that in cancer cells the apoptosis pathway is commonly altered (28)? It must also be considered that whilst during tumour establishment, autophagy maybe a mechanism through which to target tumour cells, once the tumour is established autophagy may in fact provide a way for the cancer-cells to overcome nutrient-limiting conditions especially within the internal mass of the tumour which is poorly vascularised (28, 38, 39) • Nature Reviews Drug Discovery 7, 476-477 (June 2008) • Neurodegenerative disease: A new pathway to autophagy • Charlotte Harrison • Abstract • Stimulating autophagy — a major cellular clearance route for intracellular protein aggregates — could represent a therapeutic strategy for Huntington's disease. However, the only drug that has been shown to induce autophagy in the brain is rapamycin, a mammalian target of rapamycin (mTOR) kinase inhibitor that modulates several cellular processes besides autophagy. • • • • • • • • • • • • • Neurodegenerative disorders: An aid to digestion SOURCE: home | subscribe A high-throughput screening strategy to identify novel modulators of mammalian autophagy may provide drug development leads for Huntington's disease. Autophagy, the lysosomal digestion of cytoplasmic proteins and organelles, may play a protective role in certain neurodegenerative and infectious diseases. It may also have an inhibitory role in certain cancers. Currently, the only small molecule known to regulate autophagy in mammalian brains is rapamycin. However, owing to the involvement of mTOR (mammalian target of rapamycin) proteins in many cellular processes, the long-term use of rapamycin is associated with many complications. Writing in Nature Chemical Biology, Sarkar and colleagues describe a new approach to identify novel modulators of mammalian autophagy, which might provide leads for the development of drugs for Huntington's disease. With the aim of finding safer methods of modulating autophagy, the authors performed high-throughput screening of 50,729 compounds in yeast to identify small molecules that either enhanced or suppressed the effects of rapamycin on cell growth. This resulted in the identification of a structurally non-redundant set of 21 small-molecule inhibitors of rapamycin (SMIRs) and 12 small-molecule enhancers of rapamycin (SMERs). To test the capacity of these compounds to modulate mammalian autophagy, independent of rapamycin, they assessed their ability to induce clearance of the autophagy substrate A53T -synuclein, which is associated with a form of familial Parkinson's disease. Thirteen SMIRs were shown to slow the clearance of this autophagy substrate, whereas four SMERs enhanced it. The authors then focused on a subset of autophagy-inducing SMERs that lacked toxicity in various cell lines. These might have therapeutic potential in neurodegenerative disorders such as Huntington's disease, the underlying cause of which is an expansion of a polyglutamine (polyQ) tract in the huntingtin protein. In cell models expressing the mutant huntingtin protein — another substrate for autophagy — each of the SMERs enhanced clearance of this substrate, reducing mutant protein aggregation and cell death. In a Drosophila model of Huntington's disease, the SMERs protected against neurodegeneration as assessed by the reduction in the number of visible rhabdomeres in the ommatidia of the eye over time. Interestingly, unlike rapamycin, these compounds exerted no reduction in the phosphorylation of substrates of mTOR kinase, which suggests that their induction of autophagy is mediated through an mTOR-independent mechanism, or through an unknown component of the mTOR autophagy pathway downstream of mTOR. In addition, treatment of cell models with SMERs together with rapamycin resulted in an additive effect on the clearance of A53T -synuclein and the reduction of mutant huntingtin aggregation. This study illustrates the use of a high-throughput screening strategy to identify small molecule modulators of mammalian autophagy. Structure–activity relationship analysis of the identified SMERs has highlighted functional groups that are required for their specific activity, and additional candidates for therapeutic development. Sarah Crunkhorn References Sarkar, S. et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nature Chem. Biol. 3, 331–338 (2007). | Article | PubMed | Huang, J. et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl Acad. Sci. USA 101, 16594–16599 (2004) | Article | PubMed | Rubinzstein, D. C. et al. Potential therapeutic applications of autophagy. Nature Rev. Drug Discov. 6, 304–312 (2007) | Article | PubMed | • Implication of autophagy deregulation in Parkinson's disease (PD). There are 3 types of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). Deregulation of both macroautophagy and CMA are implicated in the pathogenesis of PD. CMA is involved in the degradation of soluble wildtype alpha-synuclein, the major constituent of Lewy bodies. Nonetheless, once oligomerized or aggregated, alpha-synuclein is likely degraded by macroautophagy instead of CMA. Interestingly, A53T and A30P mutants of alpha-synuclein are poorly degraded by CMA, but are instead degraded by macroautophagy. Furthermore, the alpha-synuclein mutants inhibit CMA, reducing CMA-mediated degradation of alpha-synuclein and survival factor MEF2D. Concomitant with the inhibition of CMA, overexpression of alpha-synuclein mutants results in a compensatory activation of macroautophagy. Activation of macroautophagy is also evident in cells treated with neurotoxin MPP+, a well-established model for Parkinsonism, or following overexpression of GPR37, another protein that is present in Lewy bodies. Finally, recent studies reveal that macroautophagy also plays a role in the turnover of fragmented mitochondria. These observations highlight the potential involvement of the autophagic pathways in the pathogenesis of PD. • Nature Reviews Neurology 4, 60 (February 2008) • Rationale for treating Huntington's with a sirolimus and lithium combination • Abstract • Sarkar S et al. (2007) A rational mechanism for combination treatment of Huntington's disease using lithium and rapamycin. Hum Mol Genet 17: 170–178 PubMed • A promising treatment approach to Huntington's disease is promotion of autophagy to dispose of aggregate-prone proteins such as mutant huntingtin. Sarkar S*, Perlstein EO*, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O’Kane CJ, Schreiber SL and Rubinsztein DC. (2007) Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nature Chemical Biology 3(6):331-338. Access article Supplementary information Chemical compounds Cover highlight *Equal contribution Abstract: The target of rapamycin proteins regulate various cellular processes including autophagy, which may play a protective role in certain neurodegenerative and infectious diseases. Here we show that a primary smallmolecule screen in yeast yields novel small-molecule modulators of mammalian autophagy. We first identified new small-molecule enhancers (SMER) and inhibitors (SMIR) of the cytostatic effects of rapamycin in Saccharomyces cerevisiae. Three SMERs induced autophagy independently of rapamycin in mammalian cells, enhancing the clearance of autophagy substrates such as mutant huntingtin and A53T alpha-synuclein, which are associated with Huntington's disease and familial Parkinson's disease, respectively. These SMERs, which seem to act either independently or downstream of the target of rapamycin, attenuated mutant huntingtin-fragment toxicity in Huntington's disease cell and Drosophila melanogaster models, which suggests therapeutic potential. We also screened structural analogs of these SMERs and identified additional candidate drugs that enhanced autophagy substrate clearance. Thus, we have demonstrated proof of principle for a new approach for discovery of smallmolecule modulators of mammalian autophagy. Sarkar S, Krishna G, Imarisio S, Saiki S, O’Kane CJ and Rubinsztein DC. (2008) A rational mechanism for combination treatment of Huntington's disease using lithium and rapamycin. Human Molecular Genetics 17(2):170-178. Access article Supplementary information Abstract: Huntington's disease (HD) is caused by a polyglutamine expansion mutation in the huntingtin protein that confers a toxic gain-of-function and causes the protein to become aggregate-prone. Aggregate-prone proteins are cleared by macroautophagy, and upregulating this process by rapamycin, which inhibits the mammalian target of rapamycin (mTOR), attenuates their toxicity in various HD models. Recently we demonstrated that lithium induces mTOR-independent autophagy by inhibiting inositol monophosphatase (IMPase) and reducing inositol and IP(3) levels. Here we show that glycogen synthase kinase-3beta (GSK-3beta), another enzyme inhibited by lithium, has opposite effects. In contrast to IMPase inhibition that enhances autophagy, GSK-3beta inhibition attenuates autophagy and mutant huntingtin clearance by activating mTOR. In order to counteract the autophagy inhibitory effects of mTOR activation resulting from lithium treatment, we have used the mTOR inhibitor rapamycin in combination with lithium. This combination enhances macroautophagy by mTOR-independent (IMPase inhibition by lithium) and mTORdependent (mTOR inhibition by rapamycin) pathways. We provide proof-of-principle for this rational combination treatment approach in vivo by showing greater protection against neurodegeneration in an HD fly model with TOR inhibition and lithium, or in HD flies treated with rapamycin and lithium, compared to either pathway alone. • Sarkar S, Ravikumar B, Floto RA and Rubinsztein DC. (2009) Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death and Differentiation 16(1):46-56. Access article Abstract: The formation of intra-neuronal mutant protein aggregates is a characteristic of several human neurodegenerative disorders, like Alzheimer's disease, Parkinson's disease (PD) and polyglutamine disorders, including Huntington's disease (HD). Autophagy is a major clearance pathway for the removal of mutant huntingtin associated with HD, and many other disease-causing, cytoplasmic, aggregate-prone proteins. Autophagy is negatively regulated by the mammalian target of rapamycin (mTOR) and can be induced in all mammalian cell types by the mTOR inhibitor rapamycin. It can also be induced by a recently described cyclical mTOR-independent pathway, which has multiple drug targets, involving links between Ca2+–calpain–Gsa and cAMP–Epac–PLC-e–IP3 signalling. Both pathways enhance the clearance of mutant huntingtin fragments and attenuate polyglutamine toxicity in cell and animal models. The protective effects of rapamycin in vivo are autophagy-dependent. In Drosophila models of various diseases, the benefits of rapamycin are lost when the expression of different autophagy genes is reduced, implicating that its effects are not mediated by autophagy-independent processes (like mild translation suppression). Also, the mTOR-independent autophagy enhancers have no effects on mutant protein clearance in autophagy-deficient cells. In this review, we describe various drugs and pathways inducing autophagy, which may be potential therapeutic approaches for HD and related conditions. Sarkar S, Korolchuk V, Renna M, Winslow A and Rubinsztein DC. (2009) Methodological considerations for assessing autophagy modulators: A study with calcium phosphate precipitates. Autophagy 5(3):307-313. Access article Abstract: Autophagy has been implicated in various physiological and disease conditions in recent years. A number of small molecule modulators have been identified, both as tools and as potential therapeutics. Despite extensive characterization of autophagy in yeast, mammalian autophagy pathways are not fully understood. Recently, calcium phosphate precipitates (CPP), which are used to transfect DNA into cells, were reported to induce autophagy, when assayed up to 6 h after treatment. Because of the widespread use of this reagent, we attempted to confirm these findings. Consistent with the previous study, we showed that CPP induces autophagosome synthesis at early time-points, such as 4 h and 6 h. However, at 24 h after treatment, we were surprised to see that autophagy flux was reduced, due to impaired autophagosome-lysosome fusion. At this time point, there was an accumulation of autophagy substrates and the formation of abnormally large autophagosomes. Thus, one may need to consider assaying autophagy modulators at different time-points with a range of assays in order to understand their effects. Finally, the complex consequences of CPP on autophagy suggest that it is best avoided as a transfection reagent in studies aiming to analyse autophagy itself, or processes that are modulated by autophagy, like apoptosis. • Sarkar S, Ravikumar B and Rubinsztein DC. (2009) Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods in Enzymology Klionsky D.J., editor, Autophagy in disease and clinical applications, Academic Press 453C:83-110. Access article Abstract: Autophagy has emerged as a field of rapidly growing interest with implications in several disease conditions, such as cancer, infectious diseases, and neurodegenerative diseases. Autophagy is a major degradation pathway for aggregate-prone, intracytosolic proteins causing neurodegenerative disorders, such as Huntington's disease and forms of Parkinson's disease. Up-regulating autophagy may be a tractable therapeutic intervention for clearing these disease-causing proteins. The identification of autophagy-enhancing compounds would be beneficial not only in neurodegenerative diseasesautophagy may act as a protective pathway. Furthermore, small molecule modulators of autophagy may also be useful in dissecting pathways governing mammalian autophagy. In this chapter, we highlight assays that can be used for the identification of autophagy regulators, such as measuring the clearance of mutant aggregate-prone proteins or of autophagic flux with bafilomycin A1. Using these methods, we recently described several mTOR-independent autophagy-enhancing compounds that have protective effects in various models of Huntington's disease. • Aguado C*, Sarkar S*, Korolchuk VI, Criado O, Vernia S, Boya P, Sanz P, de Cordoba SR, Knecht E and Rubinsztein DC. (2010) Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Human Molecular Genetics 19(14):2867-2876. Access article Supplementary information Abstract: Lafora disease (LD) is an autosomal recessive, progressive myoclonus epilepsy, which is characterized by the accumulation of polyglucosan inclusion bodies, called Lafora bodies, in the cytoplasm of cells in the central nervous system and in many other organs. However, it is unclear at the moment whether Lafora bodies are the cause of the disease, or whether they are secondary consequences of a primary metabolic alteration. Here we describe that the major genetic lesion that causes LD, loss-of-function of the protein laforin, impairs autophagy. This phenomenon is confirmed in cell lines from human patients, mouse embryonic fibroblasts from laforin knockout mice and in tissues from such mice. Conversely, laforin expression stimulates autophagy. Laforin regulates autophagy via the mammalian target of rapamycin kinase-dependent pathway. The changes in autophagy mediated by laforin regulate the accumulation of diverse autophagy substrates and would be predicted to impact on the Lafora body accumulation and the cell stress seen in this disease that may eventually contribute to cell death. • • Laforin in autophagy: A possible link between carbohydrate and protein in Lafora disease? Volume 6, Issue 8 2010 Pages 1229 - 1231 Rajat Puri and Subramaniam Ganesh View affiliations Hide affiliations Rajat Puri Department of Biological Sciences and Bioengineering; Indian Institute of Technology; Kanpur, India Subramaniam Ganesh Department of Biological Sciences and Bioengineering; Indian Institute of Technology; Kanpur, India The progressive myoclonus epilepsy of Lafora disease (LD) is a fatal form of neurodegenerative disorder associated with progressive intellectual decline and ataxia in addition to epilepsy. The disease can be caused by defects in the EPM2A gene encoding laforin phosphatase or the NHLRC1 gene encoding malin ubiquitin ligase. Laforin and malin function together as a complex in the ubiquitin-proteasome system, and hence defects in proteolytic processes are thought to underlie some of the symptoms in LD. One of the pathological hallmarks of LD is the presence of cytoplasmic polyglucosan inclusions, the Lafora bodies. While Lafora bodies are known as a lesser branched form of glycogen with high phosphate content, a physiological basis for their genesis in the cytoplasm was not well understood. Recently it was shown in a mouse model for LD that loss of laforin inhibits autophagosome formation, suggesting that laforin plays a critical role in autophagosome biogenesis. The polyglucosan inclusions could be one of the substrates of autophagy, and loss of laforin might affect their sequestration into autophagosomes leading to their aggregation as Lafora bodies. Thus, laforin’s proposed role in autophagy suggests a possible link between the proteolytic system and the polyglucosan inclusions in LD. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells María Salazar, Arkaitz Carracedo, Íñigo J. Salanueva, Sonia Hernández-Tiedra, Mar Lorente, Ainara Egia, Patricia Vázquez, Cristina Blázquez, Sofía Torres, Stephane García, Jonathan Nowak, Gian María Fimia, Mauro Piacentini, Francesco Cecconi, Pier Paolo Pandolfi, Luis González-Feria, Juan L. Iovanna, Manuel Guzmán, Patricia Boya, Guillermo Velasco J Clin Invest. 2009; 119(5):1359–1372 • • THC induces autophagy via ER stress–evoked p8 and TRB3 upregulation. (A and B) Effect of ISP-1 (1 μM) on THC-induced eIF2α phosphorylation (A; 3 h; n = 3) and LC3 immunostaining (B, left panels; 18 h; percentage of cells with LC3 dots relative to the total cell number, mean ± SD; n = 3; scale bar: 20 μm) in U87MG cells. sip8, p8-selective siRNA; siTRB3, TRB3-selective siRNA. (C) Effect of THC on p8, ATF4, CHOP, and TRB3 mRNA levels of eIF2α WT and eIF2α S51A MEFs as determined by real-time quantitative PCR (8 h; n = 3). Numbers indicate the mean fold increase ± SD relative to vehicle-treated eIF2α WT MEFs. (D) Top: Analysis of p8 and TRB3 mRNA levels. Results from a representative RT-PCR experiment are shown. The numbers indicate gene expression levels as determined by real-time quantitative PCR (mean fold change ± SD relative to siC-transfected cells; n = 5). Bottom: Effect of THC on LC3 immunostaining (green) of U87MG cells transfected with siC, sip8, or siTRB3 (18 h; n = 4). The percentage of cells with LC3 dots relative to cells cotransfected with a red fluorescent control siRNA is shown in each panel (mean ± SD). Scale bar: 20 μm. (E) Effect of THC on LC3 lipidation in U87MG cells transfected with siC, sip8, or siTRB3 (18 h; n = 6). (F) Effect of THC on LC3 lipidation (top; 18 h; n = 5) and LC3 immunostaining (bottom; 18 h; percentage of cells with LC3 dots relative to the total cell number, mean ± SD; n = 4; scale bar: 40 μm) in p8+/+ or p8–/– MEFs. *P < 0.05 and **P < 0.01 compared with THC-treated U87MG (B), eIF2α WT (C), or p8+/+ (F) cells and compared with siC-transfected, THC-treated U87MG cells (D). The ubiquitin proteasome system in neuropathology by Lehman, Norman L. Acta Neuropathologica Vol. 118 Issue 3: 2009-07-27 • Cbl family E3 ligases and UBE3A E3 ligase are single proteins that associate with an E2 Ubc and the target substrate. The SCF and APC/C E3 ubiquitin ligases are multimeric complexes. The APC/C has only two known adapter subunits, Cdc20 and Cdh1 (not to be confused with E-cadherin, which is sometimes also referred to as Cdh1). The SCF can associate with several different substrate adapter proteins known as F-box proteins • The functions of neddylation are less understood; however, neddylation of cullin proteins, essential subunits of certain E3 ubiquitin ligase complexes, appears to be required for normal ligase function (Fig. 2 ). Both HECT and RING types may possess E3 ligase function as single proteins, for example the HECTtype ligase UBE3A and the RING-type ligase Cbl (Fig. 2 ). Examples include the Skp/cullin/F-box (SCF) and anaphase promoting complex/cyclosome (APC/C) E3 ubiquitin ligases that exhibit specificity for different substrates depending on the adaptor protein bound to the complex (Fig. 2 ). Familial juvenile onset Parkinson’s disease is caused by a defect in an SCF E3 ubiquitin ligase component known as parkin (PARK2) (Fig. 2 ). pVHL protein is the substrate recognition component of the ubiquitin ligase that ubiquitinates HIF-1α [ 17 ] (Fig. 2 ). The APC/C is associated with two different activator subunits that impart general substrate specificity, Cdh1 and Cdc20 (Fig. 2 ). Disorder Gene product and function Parkinson disease Autosomal dominant (early onset)α-Synuclein (SNCA) (PARK1), aggregates in Lewy bodies Autosomal dominant (late onset) Leucine-rich repeat kinase 2 (LRRK2) (PARK8), a CHIP ubiquitin ligase substrate, contains a Roc domain as found in SCF ligases Ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) (PARK5), a DUB, acts Autosomal dominant (late onset) as an E3 ligase when dimerized, polymorphisms linked to rare forms of familial disease Autosomal recessive (juvenile onset) Autosomal recessive (early onset) Autosomal recessive (early onset) Parkin (PARK2), a subunit of a SCF E3 ubiquitin ligase PINK1 (PARK6), promotes parkin ubiquitin ligase activity DJ-1 (PARK7) chaperone, promotes parkin ubiquitin ligase activity Spinocerebellar ataxias SCA1 Ataxin-1, a UBE3A E3 ligase, mutation blocks its ubiquitination and association with the ubiquitin receptor A1Up and the DUB enzyme USP7 SCA2 Ataxin-2, associates with c-Cbl E3 ligase and is involved in membrane protein endocytosis, and is a parkin E3 ligase substrate SCA3 Ataxin-3, a deubiquitinating enzyme (DUB) Prion diseases Prions may block normal function of the proteasome, HECT2D E3 ubiquitin ligase haplotypes are associated with vCJD and Kuru Autosomal recessive ALS ALS2, an endosomal membrane associated protein involved in endosome membrane fusion and trafficking, mutation decreases ALS2 protein stability Angelman syndrome Loss or mutation of UBE3A E3 ligase at Angelman/Prader–Willi locus Rett syndrome Decreased UBE3A E3 ligase due to MECP2 mutations Autism Copy number alterations of UBE3A and other UPS genes Giant axon neuropathy Mutation of gigaxonin, an E3 ubiquitin ligase IBMPFD Mutation of valosin-containing protein (VCP), involved in ubiquitinmediated processing of membrane and cytosolic proteins. Sporadic IBM VCP and ubiquitin are found in inclusion bodies. von Hippel–Lindau disease pVHL, substrate-binding subunit of ubiquitin ligase targeting HIF1-α Medulloblastoma Overexpression of several signaling pathway genes that are ubiquitinated by the SCF: c-myc, β-catenin, Gli, stabilizing mutations of Gli and β-catenin prevent their ubiquitin-dependent proteolysis Adamantinomatous craniopharyngioma Stabilizing β-catenin mutations preventing its ubiquitin-dependent proteolysis Gliomas Misregulation and mutation of cell cycle control proteins regulated by the UPS: CDKs, CDK inhibitors, p53, altered expression of APC/C E3 ubiquitin ligase regulators Emi1 and RASSF1A • • • • • 3.3 Cardiac myopathies Cardiac myocytes are terminally differentiated cells, so correct functioning of the autophagic pathway is essential for maintenance of myocyte homeostasis. Unsurprisingly, therefore, autophagic deficiencies have been associated with a variety of cardiac pathologies (reviewed in 40). One such example which has helped understanding of autophagy in the cardiac system is Danon disease. Deficiency of the autophagy gene LAMP-2 was shown to cause this disease. Patients have an increase of autophagic vacuoles leading to cardiomyopathy (15), a phenotype seen in mice with inactivation of the LAMP-2 protein (16). Whether the increase in autophagic vacuoles is due to up-regulation of autophagosome formation or due to a decrease in autophagosome – lysosome fusion is currently unclear (reviewed in 36). Autophagic vacuoles have been described in the heart tissue of patients with idiopathic dilated cardiomyopathy (41). Studies looking at models of ischaemic heart disease have also shown a role of autophagy. In chronically stunned porcine myocardium, autophagic vacuoles were seen after three episodes of ischaemia/reperfusion (I/R) and were maximal after 6 episodes, associated with increasing expression levels of autophagy-related molecules like Beclin-1. This correlated with declining levels of apoptosis between the 3-6 I/R episodes, suggesting that the autophagic response seen could be the result of a cardioprotective effect against I/R injury (42). Recently, studies investigating the role of autophagy in increasing the level of myocyte loss studied the role of Beclin-1 following a single I/R episode. These experiments showed that levels of Beclin-1 are upregulated following a single I/R episode and that this up-regulation correlates to increased autophagy. Indeed, the use of the cardioprotective agent Urocortin reduced levels of Beclin-1 and concomitantly autophagic levels. Suggesting that in order to provide protection from myocyte loss following I/R, therapies which target the autophagic response alongside the apoptotic and necrotic response should be investigated (43) • Clearly, the most fundamental question for autophagy is whether its role is harmful or protective and this remains to be answered. Indeed it would appear that autophagy could have a role during disease progression, moving from a cell protective mechanism to one contributing to cell pathology. The generation of new reagents, especially, antibodies with which to study the autophagic pathway will lead to increased understanding of the regulation of this critical homeostatic pathway. Future cellular and clinical studies will be identified which will determine whether this pathway can be manipulated to provide therapeutic potential for cardiac disease. • • • • Autophagy in heart disease By Joseph Hill * Autophagy poster and related antibodies Autophagy is an evolutionarily conserved, lysosomal pathway of engulfment, degradation and recycling of cellular contents, including long-lived proteins and organelles. This process promotes cell survival and maintains cellular homeostasis, under both resting and stress conditions. In addition, it plays a critical role in a number of clinical disorders, including heart disease. Autophagy is a complex process occurring in a stepwise manner of several stages; nucleation, expansion, and maturation/retrieval. • Williams A*, Sarkar S*, Cuddon P*, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA and Rubinsztein DC. (2008) Novel targets for Huntington's disease in an mTORindependent autophagy pathway. Nature Chemical Biology 4(5):295-305. Access article Supplementary information Chemical compounds *Equal contribution Abstract: Autophagy is a major clearance route for intracellular aggregate-prone proteins causing diseases such as Huntington's disease. Autophagy induction with the mTOR inhibitor rapamycin accelerates clearance of these toxic substrates. As rapamycin has nontrivial side effects, we screened FDA-approved drugs to identify new autophagy-inducing pathways. We found that L-type Ca2+ channel antagonists, the K+ATP channel opener minoxidil, and the Gi signaling activator clonidine induce autophagy. These drugs revealed a cyclical mTOR-independent pathway regulating autophagy, in which cAMP regulates IP3 levels, influencing calpain activity, which completes the cycle by cleaving and activating Gsa, which regulates cAMP levels. This pathway has numerous potential points where autophagy can be induced, and we provide proof of principle for therapeutic relevance in Huntington's disease using mammalian cell, fly and zebrafish models. Our data also suggest that insults that elevate intracytosolic Ca2+ (like excitotoxicity) inhibit autophagy, thus retarding clearance of aggregate-prone proteins. • Autophagy-Targeted Therapies • Autophagy is an important topic in modern cell biology. It is a dynamic process for the maintenance of cellular and metabolic homeostasis. Scientists’ interest in autophagy-related mechanisms involved in the pathogenesis of cancer and neurodegenerative diseases (Alzheimer’s disease, Huntington’s disease) and an array of other disorders is rapidly increasing. Reflecting this interest, the number of publications related to autophagy is growing exponentially. Prous Institute has initiated a research program focused on the mechanisms driving autophagy to discover new targets for the design of new small-molecule modulators of autophagy as therapeutic agents. • Several studies have shown that links may exist between autophagy and apoptosis, a subtype of programmed cell death. For this reason our Institute has a special interest in considering apoptosis in its research program Autophagy eliminates intracellular microorganisms • a | Group A Streptococci captured within an autophagosome. Image kindly provided by Tamotsu Yoshimori, Osaka University, Japan. b | Mycobacterium bovis bacillus Calmette–Guérin (BCG) present in a mycobacterial autophagosome (MAP) that is fusing with a multivesicular body (MVB). Image reproduced with permission from Ref. 42 © (2004) Cell Press. c | Herpes simplex virus type I (HSV-1) virion(s) in the process of being surrounded by an isolation membrane (left panel), engulfed inside an autophagosome (middle panel) or degraded inside an autolysosome (right panel). Image reproduced with permission from Ref. 77 © (2006) Landes Bioscience. • • • Inhibition of host autophagy by virus Autophagy is an evolutionary conserved mechanism for the sequestration and subsequent lysosomal degradation of discrete intracellular portions of eukaryotic cells, facilitating the removal of materials not degraded through the ubiquitin-proteasome pathway. In addition, autophagy plays important roles in innate and adaptive immune responses to pathogens. Several viruses have develop ways to subvert the pathway for their own benefit in order to avoid the immune response or to increase their viral replication. Beclin-1 is an essential autophagy protein and constitutes the major target for manipulation of autophagy by viruses. For example, HHV-1 ICP34.5 binds to Beclin-1 and inhibits autophagosome formation. HIV and Influenza A virus use the same mechanism through Nef and M2 respectively, that also interact with and inhibit host Beclin-1 • • • Activation of host autophagy by virus Autophagy is an evolutionary conserved mechanism for the sequestration and subsequent lysosomal degradation of discrete intracellular portions of eukaryotic cells, facilitating the removal of materials not degraded through the ubiquitin-proteasome pathway. In addition, autophagy plays important roles in innate and adaptive immune responses to pathogens Several viruses are able to activate host autophagy as a cellular survival mechanism. Indeed, viruses can activate programmed cell death during infection that prevent them from spreading to healthy tissue. By activating autophagy, viruses delay or inhibit apoptosis. For example, SV40 ST antigen protects cancer cells under glucose deprivation by triggering autophagy. KSHV Rta is able to enhance the autophagic process in order to facilitate viral lytic replication. Application of autophagy modulation to cancer therapy. From the following article: Role of autophagy in cancer Robin Mathew, Vassiliki Karantza-Wadsworth & Eileen White Nature Reviews Cancer 7, 961-967 (December 2007) • a | In apoptosis-defective tumours that are reliant on autophagy to survive metabolic stress, autophagy inhibitors can be used to induce acute necrotic cell death that may be facilitated by proteasome inhibition, enabling tumour eradication. b | In the adjuvant setting, and after elimination of a large proportion of the tumour by radiation and chemotherapy, the remaining cells can reside in a disrupted and stressed environment, susceptible to inhibition of the autophagy survival mechanism. Tumour cells in the process of metastasis can be similarly vulnerable. c | Autophagy stimulators may be therapeutically useful to either promote autophagic cell death or to prevent the damaging effects of autophagy deficiency and mismanagement of metabolic stress leading to DNA damage and tumour progression. By limiting protein, organelle and ultimately DNA damage, autophagy stimulators may suppress tumour progression. In human breast, ovarian and prostate cancers, where allelic loss of BECN1 occurs with high frequency, correction of the autophagy deficiency with autophagy stimulators may delay tumour progression by reducing the rate at which tumour-promoting mutations accumulate. Role of apoptosis and autophagy in tumorigenesis • a | Tumour-initiating mutational events such as oncogene activation promote cell proliferation, but also apoptosis, which limits tumour growth. Following the acquisition of defects in apoptosis, tumour proliferation is sustained in the absence of apoptotic cell death. b | Tumour growth is initially limited by the absence of a blood supply, which can trigger autophagy-mediated survival in the most metabolically stressed tumour regions, commonly the hypoxic center19, 20, 23. The eventual recruitment of a blood supply cures the tumour of hypoxia and metabolic stress, and the tumour cells formerly surviving through autophagy can emerge to contribute to tumour growth30. c | In tumours formed by cells with defects in both apoptosis and autophagy, necrotic cell death is stimulated in metabolically stressed tumour regions and this necrosis is associated with the activation of an inflammatory response, DNA damage and tumour progression19, 20, 23. Analogous to a wound-healing response, chronic necrosis and inflammation can stimulate angiogenesis and tumour growth24, 25, 26. The molecular regulation of autophagy. From the following article: The role of autophagy in cancer development and response to therapy Yasuko Kondo, Takao Kanzawa, Raymond Sawaya & Seiji Kondo Nature Reviews Cancer 5, 726-734 (September 2005) • In the presence of growth factors, growth factor receptor signalling activates class I phosphatidylinositol 3-phosphate kinase (PI3K) at the plasma membrane to keep cells from undergoing autophagy42. PI3K activates the downstream target AKT, leading to activation of mammalian target of rapamycin (mTOR), which results in inhibition of autophagy. p70S6 kinase (p70S6K) might be a good candidate for the control of autophagy downstream of mTOR. Overexpression of the phosphatase and tensin homologue (PTEN) gene, by an inducible promoter, antagonizes class I PI3K47 to induce autophagy. RAS has a dual effect on autophagy — when it activates class I PI3K25, autophagy is inhibited, but when it selectively activates the RAF1–mitogen-activated protein kinase kinase (MEK)–extracellular signalregulated kinase (ERK) cascade, autophagy is stimulated55. Rapamycin, an inhibitor of mTOR, induces autophagy39. A complex of class III PI3K and beclin 1 (BECN1) at the trans-Golgi network acts to induce autophagy49. This pathway is inhibited by 3methyladenine (3-MA)42. Downregulation of BCL2, or upregulation of BCL2– adenovirus E1B 19-kD-interacting protein 3 (BNIP3) or HSPIN1 at the mitochondria, also induces autophagy, indicating that BCL2 protects against autophagy52, BNIP3 (Refs 36,53,57) and HSPIN1 (Ref. 54) trigger autophagy. Autophagy is also induced by the cell death-associated protein kinase (DAPK) and the death-associated related protein kinase 1 (DRP1)51. Potential strategies for treating cancer by manipulating the autophagic process • a | Cancer cells that have defects in the autophagic pathway might be treated by replacing the autophagic signal through expression of beclin 1 (BECN1) or the phosphatase and tensin homologue (PTEN) tumour suppressor, resulting in induction of autophagy and cell death, or inhibition of proliferation. b | Cancer cells that are capable of undergoing autophagy in response to anticancer therapies might be treated with autophagy inducers, such as rapamycin, to promote autophagyinduced cell death. c | Cancer cells that undergo autophagy, to protect themselves from the effects of anticancer therapies, might be treated with autophagy inhibitors, such as bafilomycin A1 or short interfering RNAs specific for the autophagy-related genes, to induce apoptosis. A Matter of Life or Death (or Both): Understanding Autophagy in Cancer • A, in the presence of nutrients (glucose, amino acids, and growth factors), protein synthesis is stimulated and autophagy is inhibited. This is mediated through activation of mTOR (via activation of PI3K and Akt and inactivation of the tuberous sclerosis complex TSC1 and TSC2). mTOR phosphorylates S6 kinase and increases the translation of mRNAs that encode ribosomal and other proteins involved in translation. This initiates translation by phosphorylating 4EBP1, an inhibitor of initiation, causing its disassociation from eIF4E. Active eIF4 promotes cell proliferation by increasing translation of cyclin D1, c-Myc, and vascular endothelial growth factor. mTOR and S6 kinase also release the cellular check on peptide elongation by phosphorylating and inactivating eEF-2 kinase. eEF-2 kinase phosphorylates eEF-2, a 100-kDa protein that mediates the translocation step in peptide-chain elongation by inducing the transfer of peptidyl-tRNA from the ribosomal A to P site. Phosphorylation of eEF-2 at Thr56 by eEF-2 kinase decreases the affinity of the elongation factor for ribosomes and terminates elongation. Activation of TOR in yeast inhibits induction of autophagy via phosphorylation of the APG1-APG-13 complex, a process inhibited by rapamycin in both yeast and mammalian cells. B, formation of the autophagosome. This process requires the coordinated efforts of a series of gene products that respond to nutrient deprivation (autophagy regulatory complex), lipid kinase signaling molecules that participate in the formation of vesicles, ubiquitin-like proteins that complete vesicle formation, and a complex of proteins that mediate the disassembly of the autophagosome. The initial step is the envelopment of cytoplasmic materials into a phagophore or isolating membrane. This leads to the sequestration of cytoplasm into the autophagosome, which are characterized by a double membrane decorated with microtubule-associated protein 1 light chain 3 (LC3). The fusion of autophagosomes with lysosomes to for the autolysosome leads to the acidification and degradation of cytoplasmic components for recycling of amino acids and fatty acids for energy production. • • • Clin Cancer Res 2006;12:1961-1965. William N. Hait, Shengkan Jin and Jin-Ming Yang Autophagy in Cancer A Matter of Life or Death (or Both): Understanding The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells Zhen Lu, Robert Z. Luo, Yiling Lu, Xuhui Zhang, Qinghua Yu, Shilpi Khare, Seiji Kondo, Yasuko Kondo, Yinhua Yu, Gordon B. Mills, Warren S.-L. Liao, Robert C. Bast J Clin Invest. 2008; 118(12):3917–3929 • ARHI is required for autophagy. • (A–J) ARHI and rapamycin (RM) induce autophagy in ovarian cancer cells. (A–D) SKOv3-ARHI cells were transfected with GFP-LC3 and treated with or without DOX to induce ARHI expression or with or without rapamycin to inhibit mTOR activity. Scale bar: 1 μm. (E–J) ES2 and OC316 cells were not transfected or were transfected with ARHI expression vector and ARHI siRNA or control siRNA for 24 hours before they were transfected with GFP-LC3. Cells were treated with 50 nM rapamycin at the time of siRNA transfection and examined for autophagy by fluorescence microscopy 48 hours later. Scale bar: 1 μm. (K) ARHI expression is necessary for rapamycin-induced autophagy in ovarian cancer cells. ES2 and OC316 ovarian cancer cells were not transfected or were transfected with ARHI or control siRNA for 48 hours. Expression of ARHI and the control GAPDH were examined by RT-PCR. (L–U) NOSE cells undergo spontaneous autophagy. (L–O) Two NOSE cell lines were transfected with GFP-LC3 and treated with or without rapamycin. Scale bar: 1 μm. (P–U) GFP-LC3 plasmid was transfected into OSE106 cells alone or was cotransfected with ARHI siRNA or control siRNA. Transfected cells were treated with or without rapamycin. Scale bar: 1 μm. Altered capacity for apoptosis and autophagy can dictate cell fate in response to metabolic stress. • a | Apoptosis is a common response to metabolic stress in which cells activate caspases and die efficiently. For immortal epithelial cells, metabolic stress triggers apoptosis within 24 to 48 hours19, 20, 23. Execution of apoptosis occurs in less than an hour and cell viability loss is five to six orders of magnitude19. b | Defective autophagy (through loss of BECN1 or ATG5, for example) can increase apoptotic cell death in some cells in response to metabolic stress56. In human mammary cells in 3D culture in vitro, this accelerated apoptosis manifests as increased lumen formation in mammary acini20. Therefore, preservation of cell metabolism through autophagy might increase the threshold for apoptosis activation. c | In cells with defects in apoptosis, survival in metabolic stress is dependent on autophagy and is prolonged for weeks19, 20, 22, 23. During the maintenance phase, activities such as cell division and motility are sustained. Prolonged starvation and progressive autophagy causes cells to gradually shrink in size but restoration of nutrients allows recovery. In the preservation phase, cell division and motility decrease, presumably as a bioenergenic conservation effort, creating the minimal cell that is capable of recovery (MCCR). Eventually, restoration of nutrients fails to allow recovery. In this way, autophagy can be viewed as an interruptible path to cell death. d | Cells with defects in apoptosis and autophagy fail to tolerate metabolic stress, undergo metabolic catastrophe and die by necrosis36. Molecular events in autophagy. From the following article: Unveiling the roles of autophagy in innate and adaptive immunity Beth Levine & Vojo Deretic Nature Reviews Immunology 7, 767-777 (October 2007) • Autophagy is regulated by a set of autophagy-related proteins (ATG proteins). In the absence of amino acids or in response to other stimuli, ATG1 and a complex of the class III PI3K (phosphoinositide 3kinase) VPS34 and beclin 1 lead to the activation of downstream ATG factors that are involved in the initiation (a), elongation (b) and maturation (c) of autophagy. a | In amino-acid-rich conditions, VPS34 contributes to mTOR (mammalian target of rapamycin) activation and inhibition of ATG1 and autophagy. The sources of membrane for autophagosome initiation and elongation may include those containing the only known membrane integral ATG protein ATG9, redistributing between a resting location to autophagosomes in an ATG1- and PI3K-dependent manner. ATG9 redistribution may depend on ATG18, which binds phosphatidylinositol-3-phosphate (PtdIns3P). b | The elongation and shape of the autophagosome are controlled by two protein (and lipid) conjugation systems, similar to the ubiquitylation systems: the ATG12 and LC3 (also known as ATG8)–phosphatidylethanolamine (PE) conjugation pathways, which include E1-activating and E2-conjugating enzymes. ATG12 is initially conjugated to ATG7 (an E1-activating enzyme) and then is transferred to the E2-like conjugating enzyme ATG10. This intermediate presents ATG12 for conjugation to an ATG5 lysine residue. The ATG5– ATG12 conjugate, stabilized non-covalently by ATG16, triggers oligomerization on the outside membrane of the growing autophagosome, and enhances LC3 carboxy-terminal lipidation through the LC3 conjugation system. Upon autophagosome closure, ATG5–ATG12–ATG16 and LC3 (delipidated by ATG4) are recycled. c | LC3 associated with the lumenal membrane remains trapped in the autophagosome and is degraded during maturation into the autolysosome, which involves fusion of autophagosomes with late endosomes, including endosomal multivesicular bodies and lysosomal organelles, and dissolution of the internal membrane. VPS34 has a role in the formation of late endosomal multivesicular bodies and lysosomal organelles contributing to the maturation stages of autophagy. Functions of autophagy in innate and adaptive immunity during infection with intracellular pathogens. • a | Intracellular pathogens (bacteria, parasites and viruses) that are either free inside the cytosol, inside phagosomes or inside pathogencontaining vacuoles are surrounded by isolation membranes, engulfed into autophagosomes, which fuse with lysosomes, and then degraded inside autolysosomes. b | Viral nucleic acids are transferred by autophagy from the cytoplasm to intracellular compartments containing Toll-like receptor 7 (TLR7), which signals the induction of type I interferon (IFN) production. c | Viral antigens (and potentially other endogenously synthesized microbial antigens and self antigens) are engulfed into autophagosomes that fuse with MHC-class-IIcontaining late endosomes (MIICs), and then loaded onto MHC class II molecules for presentation to CD4+ T cells. Cytosolic antigens that contain a KFERQ recognition motif may also be directly imported into MIICs by chaperone-mediated autophagy. CLIP, class II-associated invariant chain peptide. Autophagy Regulators Table 1: Selected small-molecule inducers of autophagy. Treatment Target Effect Cell Line Starvation Inhibits (mTOR) Activates autophagy HeLa, HepG2, Jurkat Rapamycin Inhibits (mTOR) Activates autophagy HeLa PP242 ATP-competitive inhibitor of mTOR Activates autophagy HeLa Lithium mTOR-independent Activates autophagy HeLa Trehalose Unknown, mTOR-independent Activates autophagy HeLa Bafilomycin A1 Inhibits Vacuolar-ATPase Inhibits lysosome function HeLa Chloroquine Alkalinizes Lysosomal pH Inhibits lysosome function HeLa Tamoxifen Abolishes the inhibitory effect of PI3K Activates autophagy & inhibits lysosome function HeLa, HepG2, Jurkat Verapamil Alkalinizes Lysosomal pH Activates autophagy HeLa Hydroxychloroquine Alkalinizes Lysosomal pH Activates autophagy HeLa Loperamide mTOR-independent Activates autophagy HeLa Clonidine mTOR-independent Activates autophagy HeLa MG-132 Selective proteasome inhibitor Activates autophagy HeLa, Jurkat Norclomipramine Alkalinizes Lysosomal pH Inhibits lysosome function HeLa How to Live Long and Prosper: Autophagy, Mitochondria, and Aging • • FIGURE 1. Schematic diagram of selective and nonselective autophagy Three fundamentally different modes of autophagy are macroautophagy, microautophagy, and chaperonemediated autophagy. Depending on the specificity of the cargos, autophagy can be a selective or a nonselective process. During nonselective autophagy, a portion of the cytoplasm is sequestered into a doublemembrane autophagosome, which then fuses with the lysosome/vacuole. In contrast, the specific degradation of peroxisomes in certain conditions can be achieved by either a macro- or microautophagy-like mode, termed macropexophagy and micropexophagy, respectively. Piecemeal microautophagy of the nucleus allows the degradation of a portion of the nucleus. The specific degradation of mitochondria, termed mitophagy also takes place. A biosynthetic cytoplasm to vacuole targeting (Cvt) pathway in yeast also shares similar morphological features. Note that this schematic illustrates aspects of autophagy in both yeast cells and higher eukaryotes. Physiology, Vol. 23, No. 5, 248-262, October 2008 Int. Union Physiol. Sci./Am. Physiol. Soc. REVIEW :How to Live Long and Prosper: Autophagy, Mitochondria, and Aging Wei-Lien Yen and Daniel J. Klionsky Schematic diagram of selective and nonselective autophagy Three fundamentally different modes of autophagy are macroautophagy, microautophagy, and chaperone-mediated autophagy. Depending on the specificity of the cargos, autophagy can be a selective or a nonselective process. During nonselective autophagy, a portion of the cytoplasm is sequestered into a double-membrane autophagosome, which then fuses with the lysosome/vacuole. In contrast, the specific degradation of peroxisomes in certain conditions can be achieved by either a macro- or microautophagy-like mode, termed macropexophagy and micropexophagy, respectively. Piecemeal microautophagy of the nucleus allows the degradation of a portion of the nucleus. The specific degradation of mitochondria, termed mitophagy also takes place. A biosynthetic cytoplasm to vacuole targeting (Cvt) pathway in yeast also shares similar morphological features. Note that this schematic illustrates aspects of autophagy in both yeast cells and higher eukaryotes. Control of autophagy. Autophagy is a major housekeeping pathway and under the control of many different signaling cascades. Mammalian Target of rapamycin (mTOR) plays a central role in the regulation of autophagic activity as it integrates signaling from different sensors of cellular homeostasis. When mTOR is active in yeast it keeps an important ULK1 binding partner (ATG13) phosphorylated, thus inhibiting the induction of autophagy. While signals indicating abundant nutritional and trophic support activate mTOR (and deactivate autophagy), signals of starvation or other stressors inhibit mTOR (and activate autophagy). Autophagy can be directly stimulated by intracellular debris (such as unfolded proteins and damaged organelles) or by indicators of an overwhelmed ubiquitin-proteasome system (UPS). Also certain pathogens activate autophagy. Autophagy can be directly inhibited by genetic ablation of important Atg genes, inhibitors of the class III PI3K-complex (WM, 3-MA), high nutrient levels, and inositol signaling. More recently screenings of small compound libraries have yielded inducers and inhibitors of autophagy, both mTOR dependent and independent. And last, transcriptional regulators, such as p53, eIF2α, E2F4, or FOXO3 regulate autophagy by controlling the expression levels of many Atg genes. For further details. Jaeger and Wyss-Coray Molecular Neurodegeneration 2009 4:16 The molecular machinery of autophagy: unanswered questions Regulation of induction and vesicle nucleation. The regulation of autophagy has been characterized in studies of yeast and mammalian cells. (A) In yeast, a class III PI 3-K is required for autophagic activity and may function at the pre-autophagosomal membrane. A putative complex consisting of Atg1 kinase and several other proteins characterized as being required primarily for autophagy (in purple) or the Cvt pathway (in green) may be a downstream effector of Tor kinase to regulate the type of pathway that operates, depending on the nutritional conditions or other signals. Autophagy in yeast is primarily a starvation response; Tor, along with other regulatory components not shown (including PKA), responds to nutrient levels. In nutrient-rich conditions, Atg1 and Atg13 are more highly phosphorylated and have a lower affinity for each other; during starvation the two proteins are partially dephosphorylated. The PI 3-K complex I, consisting of Vps15, Vps34, Atg6/Vps30 and Atg14, is required for both the Cvt pathway and autophagy. (B) In mammalian cells, a class I PI 3-K is stimulated in response to the binding of a ligand to a receptor such as the insulin receptor (InR). PtdIns(3,4)P2 and PtdIns(3,4,5)P3 generated at the plasma membrane allow the binding and activation of 3-phosphoinositidedependent protein kinase 1 (PDK1) and Akt/PKB, whereas PTEN antagonizes this pathway through its 3′-phosphoinositide phosphatase activity. Akt inhibits the GTPase-activating protein complex TSC1-TSC2, resulting in the stabilization of RhebGTP, which activates Tor, resulting in the inhibition of autophagy. Both Tor and PDK1 stimulate p70S6 kinase (p70S6k). The downregulation of p70S6k activity in starvation conditions (when Tor is inhibited) might prevent excessive autophagy (Scott et al., 2004). It is also possible that p70S6k indirectly inhibits Tor by interfering with activation of the class I PI 3-K, as suggested by studies in mammalian cells (Um et al., 2004). In nutrient-rich conditions, activation of p70S6k should inhibit PI 3-K, allowing a low level of autophagy for homeostatic purposes, whereas in starvation conditions the eventual inactivation of p70S6k should allow activation of PI 3-K to prevent excessive autophagy. The class III PI 3-K serves a stimulatory role possibly similar to that of the yeast enzyme complex. The regulation of autophagy – unanswered questions • Dynamics of Atg1 complexes upon autophagy induction in different eukaryotes. (A) In yeast, under nutrient-rich conditions, the active TOR complex 1 (TORC1) hyperphosphorylates Atg13 (Kamada et al., 2010). This prevents the association of Atg1 with Atg13, which is bound to Atg17, Atg31 and Atg29, leading to inhibition of autophagy induction. Under starvation conditions when TORC1 is inactivated, Atg13 is no longer phosphorylated by TORC1, whereas Atg1 is autophosphorylated, leading to the association of Atg1 with the complex between Atg13, Atg17, Atg31 and Atg29, and subsequent autophagy induction (Cebollero and Reggiori, 2009; Chang and Neufeld, 2010; Kamada et al., 2010; Nakatogawa et al., 2009). (B) In contrast to yeast, mammalian ULK (ULK1 or ULK2, the homologs of yeast Atg1) forms a stable complex with mammalian Atg13, FIP200 (a putative counterpart of yeast Atg17) and Atg101 (an Atg13-binding protein), irrespective of TORC1 activation. Under nutrient-rich conditions, the active TORC1 associates with the ULK complex (ULK1 (or ULK2)– Atg13–FIP200-Atg101), phosphorylates ULK1 (or ULK2) and hyperphosphorylates Atg13, which inhibits the kinase activity of ULK1 (or ULK2) and thus blocks autophagy induction. Under starvation conditions when TORC1 is inactivated, TORC1 dissociates from the ULK complex, preventing phosphorylation of Atg13 and ULK1 (or ULK2) by TORC1 and leading to autophagy induction, whereas ULK1 (or ULK2) still phosphorylates Atg13 and itself, and hyperphosphorylates FIP200 (Chang and Neufeld, 2010; Mizushima, 2010; Yang and Klionsky, 2010). (C) Similar to the situation in mammals, in Drosophila Atg1 forms a complex with Atg13 irrespective of TORC1 activation (Chang and Neufeld, 2010). Under nutrient-rich conditions, the active TORC1 phosphorylates Atg13 and hyperphosphorylates Atg1, leading to the inhibition of autophagy induction. Under starvation conditions, when TORC1 is inactivated, Atg1 and Atg13 are no longer phosphorylated by TORC1, whereas Atg1 still phosphorylates itself and hyperphosphorylates Atg13, leading to autophagy induction. Figure modified from Chang and Neufeld (Chang and Neufeld, 2010) with permission. Linking different lifespan-prolonging treatments to autophagy.close • Summary of the genetic and pharmacological manipulations of autophagy that cause lifespan extension. Pharmacological treatment with spermidine, resveratrol or rapamycin, caloric restriction, depletion of p53 or overexpression of sirtuin 1 prolong lif… • • • • Can autophagy promote longevity? Frank Madeo,1 Nektarios Tavernarakis2 & Guido Kroemer3 Affiliations Journal name: Nature Cell Biology Volume: 12, Pages: 842– 846 (2010) • Abstract • Organismal lifespan can be extended by genetic manipulation of cellular processes such as histone acetylation, the insulin/IGF-1 (insulin-like growth factor 1) pathway or the p53 system. Longevity-promoting regimens, including caloric restriction and inhibition of TOR with rapamycin, resveratrol or the natural polyamine spermidine, have been associated with autophagy (a cytoprotective self-digestive process) and in some cases were reported to require autophagy for their effects. We summarize recent developments that outline these links and hypothesize that clearing cellular damage by autophagy is a common denominator of many lifespan-extending manipulations. Linking autophagy-mediated lifespan extension to cytoprotection.close • Putative mechanisms linking autophagy to the inhibition of cell death (either apoptosis or necrosis) and to the induction of longevity.