Presentation chapter 3
advertisement

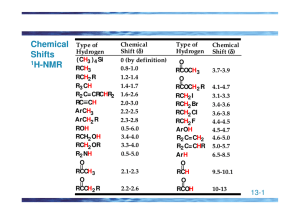
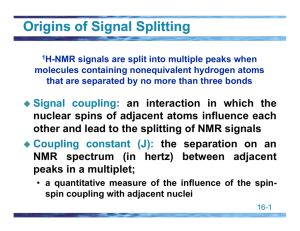
Proton NMR Spectroscopy The NMR Phenomenon • Most nuclei possess an intrinsic angular momentum, P. • Any spinning charged particle generates a magnetic field. P = [I(I+1)]1/2 h/2p where I = spin quantum # I = 0, 1/2, 1, 3/2, 2, … Which nuclei have a “spin”? • If mass # and atomic # are both even, I = 0 and the nucleus has no spin. e.g. Carbon-12, Oxygen-16 • For each nucleus with a spin, the # of allowed spin states can be quantized: • For a nucleus with I, there are 2I + 1 allowed spin states. 1H, 13C, 19F, 31P all have I = 1/2 DE = g(h/2p)Bo Spin states split in the presence of B0 -1/2 ant iparallel E +1/2 parallel no field applied field Bo When a nucleus aligned with a magnetic field, B0, absorbs radiation frequency (Rf), it can change spin orientation to a higher energy spin state. By relaxing back to the parallel (+1/2) spin state, the nucleus is said to be in resonance. Hence, NMR Presence of Magnetic Field NMR instruments typically have a constant Rf and a variable B0. A proton should absorb Rf of 60 MHz in a field of 14,093 Gauss (1.4093 T). Each unique probe nucleus (1H perhaps) will come into resonance at a slightly different and a very small percentage of - the Rf. All protons come into resonance between 0 and 12/1,000,000 (0 – 12 ppm) of the B0. • Nuclei aligned with the magnetic field are lower in energy than those aligned against the field • The nuclei aligned with the magnetic field can be flipped to align against it if the right amount of energy is added (DE) • The amount of energy required depends on the strength of the external magnetic field NMR Spectrometer • Schematic diagram of a nuclear magnetic resonance spectrometer. Energy Difference (DE) Between Two Different Spin States of a Nucleus With I=1/2 -1/2 ant iparallel E 100 MHz 200 MHz 300 MHz 400 MHz parallel +1/2 23,500 47,000 70,500 inc. magnet ic field st rengt h, Gauss B0 104,000 What Does an NMR Spectrum Tell You? • # of chemically unique H’s in the molecule # of signals • The types of H’s that are present e.g. aromatic, vinyl, aldehyde … chemical shift • The number of each chemically unique H integration • The H’s proximity to eachother spin-spin splitting Chemical Equivalence How many signals in 1H NMR spectrum? CH3 O O O O Number of Equivalent Protons CH3 1 3 5 2 O O O O 6 4 4 Homotopic H’s – Homotopic Hydrogens • Hydrogens are chemically equivalent or homotopic if replacing each one in turn by the same group would lead to an identical compound Enantiotopic H’s • If replacement of each of two hydrogens by some group leads to enantiomers, those hydrogens are enantiotopic Diastereotopic H’s • If replacement of each of two hydrogens by some group leads to diastereomers, the hydrogens are diastereotopic – Diastereotopic hydrogens have different chemical shifts and will give different signals Vinyl Protons Typical 1H NMR Scale is 0-10 ppm The d Scale Tetramethylsilane (TMS) Arbitraril y assigne d a ch e m i cal shi of d 0.00 CH3 CH3SiCH 3 CH3 TMS Chemical Shift Ranges, ppm Diamagnetic Anisotropy Shielding and Deshielding Deshielding in Alkenes Shielding in Alkynes Integration Br CH3 CH3 H H CH3 Methyl t-butyl ether (MTBE) Toluene at Higher Field Splitting patterns in aromatic groups can be confusing A monosubstituted aromatic ring can appear as an apparent singlet or a complex pattern of peaks Integral Trace Signal Splitting; the (n + 1) Rule • Peak: The units into which an NMR signal is split; doublet, triplet, quartet, multiplet, etc. • Signal splitting: Splitting of an NMR signal into a set of peaks by the influence of neighboring nonequivalent hydrogens. • (n + 1) rule: If a hydrogen has n hydrogens nonequivalent to it but equivalent among themselves on the same or adjacent atom(s), its 1H-NMR signal is split into (n + 1) peaks. Spin-Spin Splitting The Doublet in 1H NMR H a Hb C B0 C Ha i s cou ple d to bH Hb i s parall e l or an ti -parall e l 0to B Ha spli ts i n to a 1:1 doublet pe ak Hb in 1,1,2-Tribromoethane The Triplet in 1H NMR H a Hb C C Hb Ha i s cou ple d to bHan d Hb B0 Hb can both be parall e l, anti-parall e or on e paral le l an d on e an ti -paral l Ha spli ts i n to a 1:2:1 triplet pe ak Ha in 1,1,2-Tribromoethane 1,1,2-Tribromoethane The Quartet in 1HMR C H H C H H proton spl its i n to n +1 qu arte t 1:3:3:1 n = # adjacent H's B0 deshielded shielded Chemical Shift Summary of Signal Splitting • The origins of signal splitting patterns. Each arrow represents an Hb nuclear spin orientation. 1,1-Dichloroethane Ethyl benzene CH3CH2OCH3 Equivalent Protons do not Couple Pascal’s Triangle Signal Splitting (n + 1) Problem: Predict the number of 1H-NMR signals and the splitting pattern of each. O (a) CH3 CCH2 CH3 O (b) CH3 CH2 CCH2 CH3 O (c) CH3 CCH( CH3 ) 2 Differentiate using 1H NMR Methyl Isopropyl Ketone 1-Nitropropane Para Nitrotoluene Coupling Constants (J values) Bromoethane Coupling Constants – An important factor in vicinal coupling is the angle a between the C-H sigma bonds and whether or not it is fixed. – Coupling is a maximum when a is 0° and 180°; it is a minimum when a is 90°. Physical Basis for (n + 1) Rule • Coupling of nuclear spins is mediated through intervening bonds. – H atoms with more than three bonds between them generally do not exhibit coupling. – For H atoms three bonds apart, the coupling is called vicinal coupling. Physical Basis for (n + 1) Rule • Coupling that arises when Hb is split by two different nonequivalent H atoms, Ha and Hc. More Complex Splitting Patterns – Complex coupling that arises when Hb is split by Ha and two equivalent atoms Hc. More Complex Splitting Patterns – Since the angle between C-H bond determines the extent of coupling, bond rotation is a key parameter. – In molecules with free rotation about C-C sigma bonds, H atoms bonded to the same carbon in CH3 and CH2 groups are equivalent. – If there is restricted rotation, as in alkenes and cyclic structures, H atoms bonded to the same carbon may not be equivalent. – Nonequivalent H on the same carbon will couple and cause signal splitting. – This type of coupling is called geminal coupling. More Complex Splitting Patterns – In ethyl propenoate, an unsymmetrical terminal alkene, the three vinylic hydrogens are nonequivalent. More Complex Splitting Patterns – Tree diagram for the complex coupling seen for the three alkenyl H atoms in ethyl propenoate. More Complex Splitting Patterns – Cyclic structures often have restricted rotation about their C-C bonds and have constrained conformations. – As a result, two H atoms on a CH2 group can be nonequivalent, leading to complex splitting. More Complex Splitting Patterns – A tree diagram for the complex coupling seen for the vinyl group and the oxirane ring H atoms of 2methyl-2-vinyloxirane. More Complex Splitting Patterns • Complex coupling in flexible molecules. – Coupling in molecules with unrestricted bond rotation often gives only m + n + I peaks. – That is, the number of peaks for a signal is the number of adjacent hydrogens + 1, no matter how many different sets of equivalent H atoms that represents. – The explanation is that bond rotation averages the coupling constants throughout molecules with freely rotation bonds and tends to make them similar; for example in the 6- to 8-Hz range for H atoms on freely rotating sp3 hybridized C atoms. More Complex Splitting Patterns – Simplification of signal splitting occurs when coupling constants are the same. More Complex Splitting Patterns – Peak overlap occurs in the spectrum of 1-chloro-3iodopropane. – Hc should show 9 peaks, but because Jab and Jbc are so similar, only 4 + 1 = 5 peaks are distinguishable. Styrene Ha splitting in Styrene “Tree” Diagram Hc Ha C C Hb In the system below, Hb is split by two different sets of hydrogens : Ha and Hc – Theortically Hb could be split into a triplet of quartets (12 peaks) but this complexity is rarely seen in aliphatic systems Why go to a higher field strength? d0.50 d0.75 60 MHz 300 240 180 120 60 0 Hz 100 MHz 300 240 180 120 60 0 Hz 300 MHz 300 240 180 120 60 0 Hz d0.50 (t, 2H, J=10) d0.75 (t, 2H, J=10) 60 MHz 300 240 180 120 60 0 Hz 100 MHz 300 240 180 120 60 0 Hz 300 MHz 300 240 180 120 60 0 Hz Homonuclear Decoupling Allyl Bromide C H2=C HC H2Br C H2=C HC H2Br irradiate 1-Phenyl-1,2-dihydroxyethane H a Hb C C Hc OH OH 1-Phenyl-1,2-dihydroxyethane Irradiate at d 4.8