B - Computational Bioscience Program
advertisement

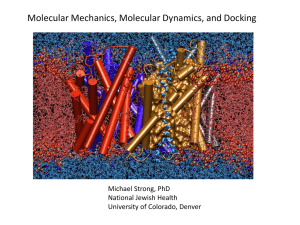
Molecular Mechanics, Molecular Dynamics, and Docking Michael Strong, PhD National Jewish Health 11/23/2010 Proteins are Dynamic Structures Aquaporin Water traveling through Aquaporin pore Control of the selectivity of the aquaporin water channel family by global orientational tuning. Tajkhorshid E, Nollert P, Jensen MØ, Miercke LJ, O'Connell J, Stroud RM, Schulten K. Science. 2002 Apr 19;296(5567):525-30. Molecular Dynamics can be used to predict protein folding (based on the physical properties of the protein) Folding proteins at x-ray resolution, showing comparison of x-ray structures (blue) and last frame of MD simulation (red): (A) simulation of villin (B) simulation of FiP35 Atomic-Level Characterization of the Structural Dynamics of Proteins Science 15 October 2010: vol. 330 no. 6002 341-346 Molecular Mechanics (MM) “The Physics of Proteins” Describe Proteins in terms of Physiochemical properties of Atoms and Bonds Calculate the dynamics of a protein, by repeated integration of the forces acting on each atom Minimum energy conformation in solution assumed to be the native state (relevant to protein folding) Molecular Mechanics •A molecule is described by interacting spheres. • Different types of spheres describe different types of atoms. • The interaction between chemically bound atoms is described by special bonding interaction terms. • The interaction of not chemically bound atoms is described by non-bonding interaction terms. • The motion of all the atoms in the molecule is described by Newtonian classical mechanics. Energy Minimization Many forces act on a protein - Hydrophobic: inside of protein avoids water - Packing: Atoms can’t be too close or too far away - Bond Angle and Length Constraints - Non-covalent (longer distance) - Hydrogen Bonds - Disulfide bonds - Ionic / Salt Bridges Can calculate all of these forces, and minimize Computationally intensive Molecular Mechanics Pros/Cons Pros: • detailed stereochemical model that describes certain aspects of biomolecules very well • conformational flexibility • dynamic model (time dependence) is possible • large systems (> 10^4 atoms) can be modeled Cons: • computationally demanding • large scale conformational changes are hard to model • no electronic (quantum) desciption, no chemical reaction (bond breaking/forming), no excited states, … • limited run times Folding@home : Distributed Computing Project Stanford University (Vijay S Pande) As of April 9, 2009 the peak speed of the project overall has reached over 5.0 native PFLOPS (8.1 x86 PFLOPS[18]) from around 400,000 active machines, including PS3. (Record) Anton massively parallel supercomputer 512-node machine: 17,000 nanoseconds of simulated time per day for a proteinwater system consisting of 23,558 atoms. In comparison, MD codes running on general-purpose parallel computers with hundreds or thousands of processor cores achieve simulation rates of up to a few hundred nanoseconds per day on the same chemical system. (enabled first microsecond MD simulation, Science 2010) (modified Amber force field) named after Anton van Leeuwenhoek : “the father of microscopy” IBM Blue Gene Popular Molecular Dynamics Programs – Linux Based AMBER (Peter Kollman, UCSF; David Case, Scripps) CHARMM (Martin Karplus, Harvard) GROMOS (Van Gunsteren, ETH, Zurich) Energy Function • Target function that MD tries to optimize • Describes the interaction energies of all atoms and molecules in the system • Always an approximation – Closer to real physics --> more realistic, more computation time (I.e. smaller time steps and more interactions increase accuracy) The energy equation (in simplistic terms) Energy = Stretching Energy + Bending Energy + Torsion Energy + Non-Bonded Interaction Energy (most computationally costly, many) These equations together with the data (parameters) required to describe the behavior of different kinds of atoms and bonds, is called a forcefield. (potential energy) The energy model • Proposed by Linus Pauling in the 1930s • Bond angles and lengths are almost always the same • Energy model broken up into two parts: – Covalent terms • Bond distances • Bond angles • Dihedral angles • Non-covalent terms • Forces at a distance between all non-bonded atoms Bond length • Spring-like term for energy based on distance • kb is the spring constant of the bond. r0 is the bond length at equilibrium. Unique kb and r0 assigned for each bond pair, i.e. C-C, O-H Bond bend k is the spring constant of the bend. 0 is the bond length at equilibrium. Unique parameters for angle bending are assigned to each bonded triplet of atoms based on their types (e.g. C-C-C, C-O-C, CC-H, etc.) Torsion Energy Energy needed to rotate about bonds. Only relevant to single bonds A controls the amplitude of the curve n controls its periodicity shifts the entire curve along the rotation angle axis (). The parameters are determined from curve fitting. Unique parameters for torsional rotation are assigned to each bonded quartet of atoms based on their types (e.g. C-C-CC, C-O-C-N, H-C-C-H, etc.) Non-bonded Energy Van der Waals – preferred distance between atoms If atoms are polar, some will have partial electrostatic charges (attract if opposite, repel if same) A and B constants depending on atom type. A determines the degree the attractiveness A determines the degree the attractiveness B determines the degree of repulsion B determines the degree of repulsion q is the charge q is the partial atomic charge Why simulate motion? • • • • Predict structure Understand interactions Understand properties Experiment on what cannot be studied experimentally Energy minimization • Given some energy function and initial conditions, we want to find the minimum energy conformation. (steepest decent algorithm) • Various programs: Charmm, Amber are two most widely used (and packaged), DE Shaw’s Desmond Folding proteins at x-ray resolution, showing comparison of x-ray structures (blue) and last frame of MD simulation (red): (A) simulation of villin (B) simulation of FiP35 Atomic-Level Characterization of the Structural Dynamics of Proteins Science 15 October 2010: vol. 330 no. 6002 341-346 • Solvation models: water & salt are very important to molecular behavior. Must model as many water atoms as protein atoms (often more than molecule, explicit model). Molecular Dynamics • Molecules, especially proteins, are not static. – Dynamics can be important to function – Molecules allowed to interact for a period of time (fs steps) – Consider number of particles, timestep, total time duration, nanoseconds to microseconds (several CPU days to CPU years) (nanosecond simulation -> millions of calculations) – 10usec simulation -> 3 months • Trajectories, not just minimum energy state. – MM ignores kinetic energy, does only potential energy – MD takes same force model, but calculates F=ma and calculates velocities of all atoms (as well as positions) Docking • Computation to assess binding affinity • Looks for conformational and electrostatic "fit" between proteins and other molecules • Optimization again: what position and orientation of the two molecules minimizes energy? • Large computations, since there are many possible positions to check, and the energy for each position may involve many atoms Docking Similar equation A and B constants depending on atom type. A determines the degree the attractiveness B determines the degree of repulsion q is the partial atomic charge Molecular Docking Start with PDB file, homology model, etc Add Hydrogens Select Grid Box Identify molecule to be docked >10 runs, > 1 million evaluations Genetic Algorithm Molecular Docking (Example in TB) A B C Heme W107 H108 W321 Isoniazid KatG Dimer with 2 heme molecules R104 T314 S315 KatG Heme Binding Site is also the site of Isoniazid Activation D P136 A139 L205 P232 T314 D282 S315 A281 G316 I317 Isoniazid Docked into the KatG active site Steps: 1. Get crystal structure of protein from PDB 2. Get small molecule coordinates (DrugBank) 3. Use AutoDock 4. Add Hydrogens to both structures 5. Identify potential binding site, specify GridBox (center on heme) (dimensions 40x40x40) 6. Dock using Genetic Algorithm, 10 runs, 2,500,000 evaluations Virtual Screening • Docking small ligands to proteins is a way to find potential drugs. Libraries • A small region of interest (pharmacophore) can be identified, reducing computation • Empirical scoring functions are not universal • Various search methods: – Rigid provides score for whole ligand (accurate) – Flexible breaks ligands into pieces and docks them individually Docking example Biotin docking with Streptavidin, from Olsen lab at Scripps Macromolecular docking • Docking of proteins to proteins or to DNA • Important to understanding macromolecular recognition, genetic regulation, etc. • Conceptually similar to small molecule docking, but practically much more difficult – Score function can't realistically compute energies – Use either shape complementarity alone or some kind of mean field approximation Docking Resources • AutoDock http://autodoc.scripps.edu/ • Dock http://www.cmpharm.ucsf.edu/kuntz/dock.html • Movie: Docking