Presentation - Oklahoma State University
advertisement

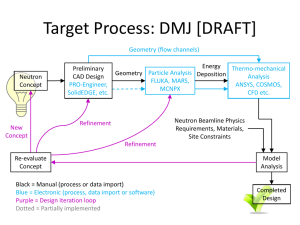
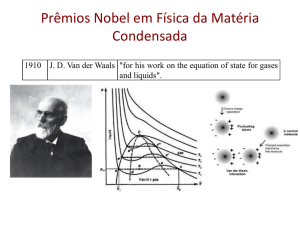
Neutron Diffraction and Scattering in Biology Penghui Lin Department of Physics Oklahoma State University 10/4/2013 Neutron Scattering • Neutron Reflection (Neutron Reflectometry) • Small Angle Neutron Scattering • Neutron diffraction (Neutron Crystallography) • Spectroscopy and Imaging Structure determination • X-ray diffraction----spatial distribution of electrons • Electron diffraction----Coulomb forces • Neutron diffraction---strong nuclear forces X-ray • NMR • IR NMR Electron Microscopy Hybrid Other X-ray vs Neutron X-ray v.s. Neutron Crystallography Crystal -> Diffraction pattern -> Electron density -> Model Crystal -> Diffraction pattern -> Nuclear density -> Model X-ray v.s. Neutron Information from NC • Equivalence: neutron scattering not strongly depends on Z (especially for hydrogen detection which X-ray or electron diffraction can not see) • Clearly distinguish between neighboring atoms. (For biology, particularly N, C and O) • • • • Contrast between H and D Locate the solvent orientation around protein Thermal motions of hydrogen containing groups Weak interaction with materials, deep penetration and non-destructive Crucial Hydrogen • Dominance of H2O molecules in living cells • Hydrogen bonds provide stability and versatility for biological macromolecules • Proton transfer and exchange is critical in many reactions • Hydration and protonation states are important Neutron diffraction in structural studies • • • • Location of Hydrogen atoms Solvent Structure Hydrogen exchange Low resolution studies H D % bc bi 99.985 -3.741 25.27 0.015 6.671 4.04 sc 1.758 5.592 si 80.27 2.051 ss 82.03 7.643 sa 0.3326 0.000519 Bound atom scattering length Neutron sees Fissure Reactor Chained reactions Continuous flow 1 neutron per fission 180 MeV neutron 1015/cm2/s Spallation source No chain reaction Pulsed 40 neutrons per proton 30 MeV neutron 1016/cm2/s Neutron source Neutron source Combined with Fourier Transform Main problem • Low flux of neutron beams • Structures are large while scatterings are weak, so large single crystals are required, 1 mm3 is the limit due to the reasonable data collection time of 10-14 days per data set • Hydrogen produces a high level background (80 barn scattering factor) Solution • Broad bandpass (maximize the neutron flux and the reflections on the detector) • Cylindrical neutron image plate (LADI at ILL has a solid angle >2π) • Isotope substitution of D to H Developments • In reactors: – Neutron image plates – Quasi-Laue methods • In spallation: – Time of flight Laue method – Electronic detectors • New facilities and methods for sample perdeuteration and crystallization • New approaches and computational tools for structure determination New neutron sources Applications EXAMPLE I D-XYLOSE ISOMERASE (XI) XI: Xylose Isomerase Mechanism of Aldo to Keto Environment OD- in XI-xylulose D2O in native XI M1: structural metal M2: catalytic metal Kovalevsky 2008 Biochemistry Active site of XI-xylulose Doubly protonated singly protonated Kovalevsky 2008 Biochemistry Kovalevsky 2008 Biochemistry Applications EXAMPLE II CRYO COOL CONCANAVALIN A Concanavalin A Saccharide-binding protein Legume lectin family Extensice β-sheet arrangement Two metal binding sites PDB: 3CNA Waters in the saccharide-binding site 293K Habash 2000 Acta Crystallogr D Biol Crystallogr. 15K Blakeley 2004 PNAS H-bond network Blakeley 2004 PNAS Water comparison Compare to room temperature NC 15K 227 water sites are identified with 19.2 Å2 B factor 167 are D2O with 17.6 Å2 B factor 60 are OD- or oxygen atoms with 32.2 Å2 B factor 293K 148 water sites are identified with 43 Å2 B factor 88 are D2O with 37.8 Å2 B factor 60 are OD- or oxygen atoms with 50.6 Å2 B factor Compare to low temperature (100K) XC Among the 16 buried waters, 9 matched the positions in the X-ray structure (56.3%) Among the 211 surface waters, 35 matched the positions in the X-ray structure (16.6%) Blakeley 2004 PNAS Conserved water molecules W1 W6 W75 Neutron 15K Neutron 293K X-ray 110K Only 22 water molecules are conserved in positions Blakeley 2004 PNAS Applications EXAMPLE III SANS IN LIPID UNIFORMITY Lipid raft Proposal: Hybrid lipids align in a preferred orientation at the boundary of ordered and disordered phases, lowering the interfacial energy and reducing domain size Fluorescence microscopy of GUVs ρ ≡ χDOPC/(χDLPC+χDOPC) FRET and SANS results Small Angle Neutron Scattering Förster Resonance Energy Transfer Conclusion • A complementary technique to others • Sensitive to light atoms, especially hydrogen • Can be applied to various materials References 1. Heberle FA, et al. (2013) Hybrid and Nonhybrid Lipids Exert Common Effects on Membrane Raft Size and Morphology. Journal of the American Chemical Society. 2. Comoletti D, et al. (2007) Synaptic arrangement of the neuroligin/beta-neurexin complex revealed by X-ray and neutron scattering. Structure 15(6):693-705. 3. Stuhrmann HB (2004) Unique aspects of neutron scattering for the study of biological systems. Rep Prog Phys 67(7):1073-1115. 4. Habash J, et al. (2000) Direct determination of the positions of the deuterium atoms of the bound water in concanavalin A by neutron Laue crystallography. Acta Crystallogr D 56:541-550. 5. Holt SA, et al. (2009) An ion-channel-containing model membrane: structural determination by magnetic contrast neutron reflectometry. Soft Matter 5(13):2576-2586. 6. Blakeley MP, Langan P, Niimura N, & Podjarny A (2008) Neutron crystallography: opportunities, challenges, and limitations. Curr Opin Struc Biol 18(5):593-600. 7. Niimura N, Chatake T, Ostermann A, Kurihara K, & Tanaka I (2003) High resolution neutron protein crystallography. Hydrogen and hydration in proteins. Z Kristallogr 218(2):96-107. 8. Collyer CA & Blow DM (1990) Observations of Reaction Intermediates and the Mechanism of Aldose-Ketose Interconversion by DXylose Isomerase. Proceedings of the National Academy of Sciences of the United States of America 87(4):1362-1366. 9. Blakeley MP, Kalb AJ, Helliwell JR, & Myles DAA (2004) The 15-K neutron structure of saccharide-free concanavalin A. Proceedings of the National Academy of Sciences of the United States of America 101(47):16405-16410. 10. Blakeley MP, et al. (2008) Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase. Proceedings of the National Academy of Sciences of the United States of America 105(6):1844-1848. 11. Lakey JH (2009) Neutrons for biologists: a beginner's guide, or why you should consider using neutrons. J R Soc Interface 6:S567S573. 12. Kovalevsky AY, et al. (2008) Hydrogen location in stages of an enzyme-catalyzed reaction: Time-of-flight neutron structure of Dxylose isomerase with bound D-xylulose. Biochemistry-Us 47(29):7595-7597. Thanks