Modeling
advertisement
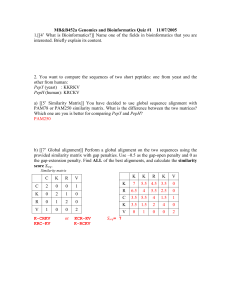
Bioinformatics how to … use publicly available free tools to predict protein structure by comparative modeling Proteins are 3D objects with complex shapes Over 60,000 protein structures have been determined, mostly by X-ray crystallography (PDB) 3D structure of ~70% of bacterial and 50% of human proteins can be predicted (comparative modeling) A predicted model simply illustrates our assumptions No assumptions, this is nature telling us how it is GNAAAAKKGSEQESVKEFLAKAKEDFLKKWENPA QNTAHLDQFERIKTLGTGSFGRVMLVKHKETGNH FAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPF LVKLEYSFKDNSNLYMVMEYVPGGEMFSHLRRIG RFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPE NLLIDQQGYIQVTDFGFAKRVKGRTWTLCGTPEY LAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPF FADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNL LQVDLTKRFGNLKDGVNDIKNHKWFATTDWIAIY QRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSIN EKCGKEFSEF Sequence Assumption (protein A is Similar to protein B) Result (protein A is Similar to protein B) How do we know that these proteins are similar? Well studied protein Unknown protein GLLTTKFVSLLQEAKDGVLDLKL AADTLAVRQKRRIYDITNVLEGIG similarity LIEKKSKNSIQW prediction SRRSASHPTYSEMIAAAIRAEKS RGGSSRQSIQKYIKSHYKVGHN ADLQIKLSIRRLLAA How can we make such assumptions? Statistical reliability of the prediction E-value - the number of hits one can "expect" to see just by chance when searching a database of a particular size (closer to zero the better) Z-score – score expressed as a distance from the mean calculated in standard deviations (the bigger the better) Similar, but not homologous phosphoribosyltransferase and viral coat protein, identity: 42%, different folds, different functions . . . . . 99 IRLKSYCNDQSTGDIKVIGGDDLSTLTGKNVLIVEDIIDTGKTMQTLLSLVRQY.NPKMVKVASLLVKRTPRSVGY 173 : ||. ||| || |. || | : | | | | || | || |:| | ||.| | 214 VPLKTDANDQ.IGDSLY....SAMTVDDFGVLAVRVVNDHNPTKVT..SKVRIYMKPKHVRV...WCPRPPRAVPY 279 Different, but homologous Histone H5 and transcription factor E2F4, identity 7%, similar fold, similar function (DNA binding) PTYSEMIAAAIRAEKSRGGSSRQSIQKYIKSHYKVGHNADLQIKLSIRRLLAAGVLKQTKGVGASGSFRL | | | | | GLLTTKFVSLLQEAKD-GVLDLKLAADTLA------VRQKRRIYDITNVLEGIGLIEKKS----KNSIQW Steps in comparative modeling Recognition Are there any well characterized proteins similar to my protein? Alignment What is the position-by-position target/template equivalence Modeling Model analysis What is the detailed 3D structure of my proteins Is my model any good? Recognition BLAST, PSI-BLAST or PFAM, FFAS, metaserver (bioinfo) Name (PDB code) of the template Statistical significance of the match (Zscore, e.value, p.value, points) Alignment The same tools as in recognition (perhaps with different parameters), editing by hand Position by position equivalence table Modeling Commercial programs Accelrys (Insight) Tripos (Sybyl) … Freeware/shareware /servers Modeller (Andrej Sali) Jackal (Barry Honig) SCRWL (Roland Dunbrack) SwissModel Model quality Empirical energy based tools PSQS (http://www1.jcsg.org/psqs/psqs.cgi) SwissPDB viewer Geometric quality Procheck, SFCHECK, etc. (http://www.jcsg.org/scripts/prod/validatio n/sv3.cgi) Expectations of comparative modeling 75 50 25 0 Easy – 100-40% sequence id - strong sequence similarity, strong structure similarity, obvious function analogy Difficult – 40%-25% - twilight zone sequence similarity, increasing structure divergence, function diversification Fold prediction – below 25% seq id. no apparent sequence similarity extreme function divergence Challenges of comparative modeling Modeling Recognition Alignment Trivial Trivial Simple Loop modeling Trivial Easy Simple Loop modeling Simple Challenging Challenging Difficult Very difficult Significant errors Often impossible Significant errors Often impossible Alignment, backbone shifts Alignment, backbone shifts Recognition Challenges 100 80 60 40 20 Hands-on Activity Click below for a hands-on, “bioinformatics how to” activity Go to http://bioinformatics.burnham.org/ Click Structure Biology Course - “Protein homepage. OR Go to…. Modeling Tutorial” Link in the http://bioinformatics.burnham.org/SSBC/modeling.html