Introduction to ARB - Microbial Genome Program
advertisement
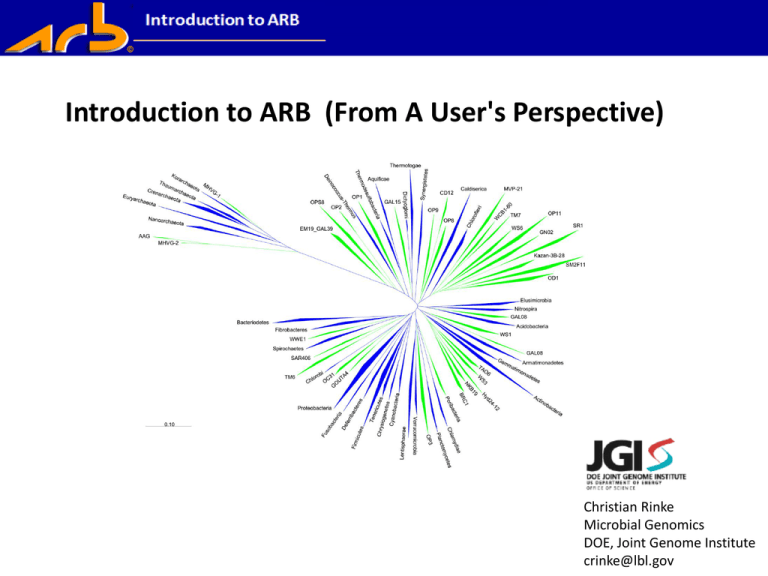
Introduction to ARB (From A User's Perspective) Christian Rinke Microbial Genomics DOE, Joint Genome Institute crinke@lbl.gov Reasons for using molecular markers Sort and classify organisms Unravel evolutionary relationships Track back the origin of organisms Problems when using molecular markers for Phylogeny Lateral gene transfer/ Genome plasticity Orthologous/paralogous genes Examples of Phylogenetic Markers • • • • • • • • • 16S rRNA 23S rRNA Elongationfactors (EF-Tu; EF-G) ATP-Synthase Reg A Hsp60 RNA-Polymerase Gyrase other housekeeping genes • Presence in all organisms • Functional constancy • Comprehensive database rRNA as Phylogenetic Marker 21 proteins 16S rRNA 30S 70S Ribosome subunits 50S 34 proteins 5S rRNA 23S rRNA Escherichia coli 16S rRNA Primary and Secondary Structure rRNA as Phylogenetic Marker Advantages • Functional constancy • Ubiquitous distribution • Relatively large size (information content) • Conserved and highly variable structural elements • Almost no lateral gene transfer Drawbacks • No continuous sequence change • Multiple genes/operons • Different species with identical 16S rRNAs ARB: a software environment for sequence data • ARB (ARBor, Latin: tree) • UNIX based – but graphically oriented package • Toolbox for analyzing sequence data, emphasis on phylogeny reconstruction. • Developed by: Ludwig et al. (2004) ARB: a software environment for sequence data. Nucleic Acids Research. 32(4):1363-1371. doi:10.1093/nar/gkh293 Why ARB Introduction to ARB Why ARB (+) Handles large databases (+) Comprehensive package: sequence alignment, meta data, filter calculation, phylogenetic analyses, FISH probe design (+) available at no cost : www.arb-home.de. (+) ready made databases available (+/-) Unix based - SuSE Linux (Ubuntu Linux, Mac OSX) (installation rather straight forward); ribocon (+/-) Under development/ supporting information Live Demo: 16S rRNA sequence of unknown organisms Strategy: -- Retrieve related reference sequences from Downloadclosely database Genbank via BLAST > build reference data set - Compile sequences in Fasta format in one text file - Perform sequence alignment - Using tree reconstruction to dentify organism Introduction to ARB ARB database >>> SILVA rRNA database project: http://www.arb-silva.de/download/arb-files/ • quality checked and regularly updated • aligned • small (16S/18S, SSU) and large subunit (23S/28S, LSU) ribosomal RNA (rRNA) sequences • all three domains of life Pruesse, E., C. Quast, K. Knittel, B. Fuchs, W. Ludwig, J. Peplies, and F. O. Glöckner. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nuc. Acids Res. 2007; Vol. 35, No. 21, p. 7188-7196 other sources: >>> greengenes (greengenes.lbl.gov) >>> Ribosomal Database Project (rdp.cme.msu.edu) Live Demo Phylogenetic tree reconstruction Secondary structure helix symbols in ARB editor Phylogenetic tree reconstruction Phylogenetic tree reconstruction Algorithmic (distance methods) Neighbor Joining Tree-searching (character based methods) Parsimony Maximum Likelihood Bayesian analysis Phylogenetic ARB is useful for nextgen amplicon sequencestree reconstruction Pre-sequencing: • Validating primer specificity • Primer design Data QC: • Viewing sequence alignments and checking taxonomic identifications • Resolving ‘unknowns’ • Evaluating conserved & variable gene regions Post-processing: • Comparative phylogenetic context • Specific probe or primer design Help http://www.arb-home.de/documentation.html http://help.arb-home.de/ http://tech.groups.yahoo.com/group/arb_users/ Ribocon: http://www.ribocon.com/home/arb/ – ARB instillation DVD – Workshops Introduction to ARB Introduction to ARB Introduction to ARB (From A User's Perspective) Christian Rinke Microbial Genomics DOE, Joint Genome Institute crinke@lbl.gov