Presentation
advertisement

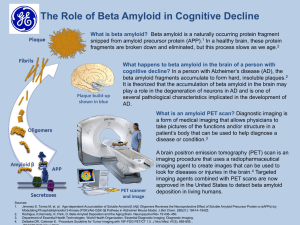
KINETICS OF THE PRION PROTEIN: STRUCTURE, MISFOLDING, DISEASE, AND STABILITY Rhiannon Aguilar HONR299J Final Presentation Spring 2014 BACKGROUND ON THE PRION “Proteinaceous Infectious Particle” Amyloid disease: visible protein deposits that can be stained Stanley Prusiner, 1982 Plaques found in 10% of CJD, higher percentage in other TSEs Two distinct forms, PrP-C and PrR-res Membrane-bound protein, 254 amino acids, 2 glycosylation sites Conserved between species, but with slight changes resulting in a disease species barrier PrP 27-30, fragment created by digestion, can form amyloid Highly expressed in CNS, lymphatic tissue HELICES AND PLEATED SHEETS α-Helix: 3.6 amino acids/turn, righthanded spiral β-pleated sheet: parallel or anti-parallel sheets with a kinked shape, connected by a loop http://www.mun.ca/biology/scarr/MGA2_0318b.html PRION STRUCTURE Cellular PrP Lots of alpha helices Point mutations can cause slight changes in structure that make misfolding favorable Biologically interesting fragment: 108-218 Huang, Prusiner, and Cohen 1996 MISFOLDED STRUCTURE Predominantly beta-pleated sheets Presumably, this structure is more likely to form aggregates Same biologically interesting fragment (108218) Huang, Prusiner, and Cohen 1996 DISEASE MECHANISM: REFOLDING VS SEEDING Refolding: Conversion is very slow normally Misfolded protein acts as enzyme to re-fold normal Seeding: Conversion is in constant equilibrium Seeds form when Sc form accumulates, prevents return to normal state http://www.nature.com/nri/journal/v4/n9/images/nri14 37-f1.jpg ANIMATION OF “REFOLDING” MODEL http://learn.genetics.utah.edu/content/molecu les/prions/ (Slide 7) PRP DIMERIZATION PrP can form a covalent dimer Third helix swaps position to form a covalent bond with a second molecule Forms a β-sheet at the interface Possibly a precursor to aggregation in disease http://www.nature.com/nsmb/journal/v8/n9/full/nsb0901770.html MORE PICTURES OF DIMERIZATION Top: Green/Pink are the two halves of the dimer, Blue is the monomer superimposed Bottom: Left is the two halves of the dimer, pulled apart, and right is two monomers http://www.nature.com/nsmb/journal/v8/n9/full/nsb0901770.html SIGNIFICANCE OF THIS DIMERIZATION? 17 amino acids present in familiar SE’s are located on the flipped helix Mutations may make this flipping easier, facilitate protein conformational change Covalent dimers present in hamster scrapie brains Formation of new covalent linkages = protein unfolding/refolding Red: Amino acids mutated in familial SE’s Bottom left: Met129, site of the Val mutation that is a marker for CJD http://www.nature.com/nsmb/journal/v8/n9/full/nsb090 1-770.html FOLDING KINETICS Fast-folding Easily folds incorrectly Has mutations which perturb folding but do not change stability Important “nucleus” located between helices 2 and 3 (3rd helix is moved in dimer formation) KINETICS: EFFECT OF TEMPERATURE ON STABILITY Folded + GnHCL Unfolded Plot relates to reverse reaction (Unfolded Folded) Native protein is most stable at ~285K (11.85°C) http://www.pnas.org/content/106/14/5651.full KINETICS: FOLDING OF NATIVE PRP J. Biol. Chem. (2002) Mutate Trp to Phe gives fluorescence to folded protein Experiments done at 5°C b/c too fast at 25°C Results: Prion folds/unfolds with a kinetic intermediate First conclusive evidence for a folding intermediate Prev. results say that mouse PrP does not have an intermediate Possible reason for species barrier? KINETICS: EFFECT OF MUTATIONS Folding/unfolding goes through a partially-folded intermediate state In most familial mutant versions, the intermediate is extra stable Intermediate is highly stabilized by 7/9 mutations The intermediate is likely to aggregate Native protein needs PrP-res seed, but maybe mutant intermediate states can aggregate on their own? http://www.jbc.org/content/279/17/18008.long STABILITY OF AGGREGATES: AMYLOID STABILITY Amyloid analogue synthesized: KFFEAAAKKFFE Aromatic pi-pi stacking (phenylalanine) Charge attraction Β-sheet interactions similar to silk http://www.pnas.org/content/102/2/315.full http://www.pearsonhighered.co m/mathews/ch06/fi6p12.htm STRUCTURAL STABILITY OF PRION AGGREGATES Differences in structure at aggregate core results in differential stability Less stable = shorter incubation time (Prusiner) Synthesize PrP aggregates in two conditions: 2M GnHCl and 4M GnHCl Produces 2 different stabilities when denaturation is attempted Open circles: 2M, Closed circles: 4M 4M shows significantly higher stability http://www.jbc.org/content/289/ 5/2643.long STRUCTURAL STABILITY OF PRION AGGREGATES Differential stability seems to only relate to packing arrangement, not the protein secondary structure Tighter packing = protease resistance? Stability of disease amyloid may relate to conformation of amyloid innoculated http://www.jbc.org/content/289/5/264 3.long THERMODYNAMIC STABILITY: STUDIES OF INSULIN AMYLOID Thermal decomposition results in loss of mass and release of gas Occurs at lower temperature for native molecule than for amyloid http://www.plosone.org/article/info%3Adoi% 2F10.1371%2Fjournal.pone.0086320 THERMODYNAMIC STABILITY: STUDIES OF INSULIN AMYLOID http://www.plosone.org/article/ info%3Adoi%2F10.1371%2Fjou rnal.pone.0086320 Incubation at increasing temperatures decomposes fibril structure Incubation at 100°C shows little effect on structure (if anything, may be more stable?) Might be unfolding/refolding to a more stable structure? Autoclaves at 120-130°C not sufficient! Higher temperatures seem very effective