Applying Linked Data Principles to Represent Patient`s Electronic
advertisement
Applying Linked Data Principles to Represent Patient’s Electronic Health Records
at Mayo Clinic: A Case Report
Jyotishman Pathak, PhD Richard C. Kiefer Christopher G. Chute, MD, DrPH
Division of Biomedical Statistics and Informatics, Department of Health Sciences Research
Mayo Clinic, Rochester, MN
Background and Aims
Patient recruitment is a huge bottleneck in
conducting clinical trials and research studies
50% of time is spent in recruitment
Low participant rates (~5%)
Clinicians lack resources to help find patients
appropriate studies; patients encounter
difficulties locating appropriate studies
Semantic Web
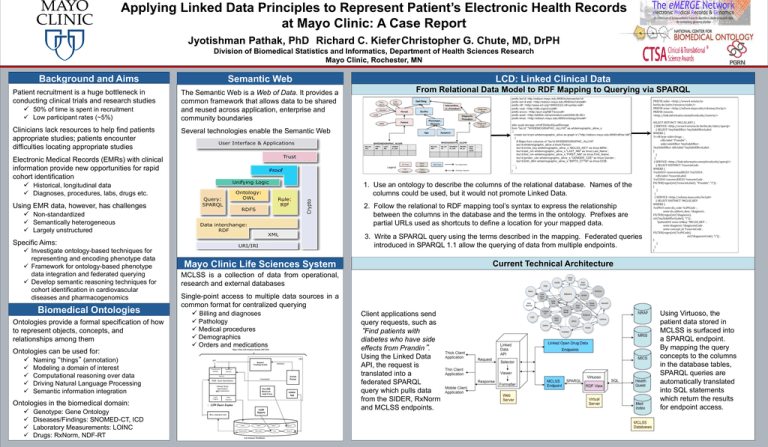
LCD: Linked Clinical Data
The Semantic Web is a Web of Data. It provides a
common framework that allows data to be shared
and reused across application, enterprise and
community boundaries
From Relational Data Model to RDF Mapping to Querying via SPARQL
Several technologies enable the Semantic Web
Electronic Medical Records (EMRs) with clinical
information provide new opportunities for rapid
cohort identification
Historical, longitudinal data
1. Use an ontology to describe the columns of the relational database. Names of the
columns could be used, but it would not promote Linked Data.
Diagnoses, procedures, labs, drugs etc.
2. Follow the relational to RDF mapping tool’s syntax to express the relationship
between the columns in the database and the terms in the ontology. Prefixes are
partial URLs used as shortcuts to define a location for your mapped data.
Using EMR data, however, has challenges
Non-standardized
Semantically heterogeneous
Largely unstructured
3. Write a SPARQL query using the terms described in the mapping. Federated queries
introduced in SPARQL 1.1 allow the querying of data from multiple endpoints.
Specific Aims:
Investigate ontology-based techniques for
representing and encoding phenotype data
Framework for ontology-based phenotype
data integration and federated querying
Develop semantic reasoning techniques for
cohort identification in cardiovascular
diseases and pharmacogenomics
Biomedical Ontologies
Ontologies provide a formal specification of how
to represent objects, concepts, and
relationships among them
Ontologies can be used for:
Naming “things” (annotation)
Modeling a domain of interest
Computational reasoning over data
Driving Natural Language Processing
Semantic information integration
Ontologies in the biomedical domain:
Genotype: Gene Ontology
Diseases/Findings: SNOMED-CT, ICD
Laboratory Measurements: LOINC
Drugs: RxNorm, NDF-RT
PREFIX sider: <http://www4.wiwiss.fuberlin.de/sider/resource/sider/>
PREFIX semr: <http://edison.mayo.edu/schemas/lss1p/>
PREFIX rxnorm:
<http://link.informatics.stonybrook.edu/rxnorm/>
SELECT DISTINCT ?MCLSS_KEY {
{ SERVICE <http://www4.wiwiss.fu-berlin.de/sider/sparql>
{ SELECT ?mySideEffect ?mySideEffectLabel
WHERE {
?x rdf:type sider:drugs ;
rdfs:label "Prandin" ;
sider:sideEffect ?mySideEffect .
?mySideEffect rdfs:label ?mySideEffectLabel .
}
}
}
{ SERVICE <http://link.informatics.stonybrook.edu/sparql/>
{ SELECT DISTINCT ?rxnormCode
WHERE {
?rxAUIUrl rxnorm:hasRXCUI ?rxCUIUrl .
rdfs:label ?rxnormLabel
?rxCUIUrl rxnorm:RXCUI ?rxnormCode .
FILTER(regex(str(?rxnormLabel), "Prandin", "i")) .
}
}
}
{ SERVICE <http://edison.mayo.edu/lss1p#>
{ SELECT DISTINCT ?MCLSS_KEY
WHERE {
?icd9Url semr:dx_code ?icd9Code ;
semr:dx_abbrev_desc ?diagnosis .
FILTER(regex(str(?diagnosis),
str(?mySideEffectLabel), "i")) .
?patientUrl semr:whkey ?MCLSS_KEY ;
semr:diagnosis ?diagnosisCode .
semr:concept_id ?rxnormCode .
FILTER(regex(str(?icd9Code),
str(?diagnosisCode), "i")) .
}
}
}
}
Current Technical Architecture
Mayo Clinic Life Sciences System
MCLSS is a collection of data from operational,
research and external databases
Single-point access to multiple data sources in a
common format for centralized querying
Billing and diagnoses
Pathology
Medical procedures
Demographics
Orders and medications
Client applications send
query requests, such as
“Find patients with
diabetes who have side
effects from Prandin”.
Using the Linked Data
API, the request is
translated into a
federated SPARQL
query which pulls data
from the SIDER, RxNorm
and MCLSS endpoints.
+
Using Virtuoso, the
patient data stored in
MCLSS is surfaced into
a SPARQL endpoint.
By mapping the query
concepts to the columns
in the database tables,
SPARQL queries are
automatically translated
into SQL statements
Personnel
which return the results
for endpoint access.