Luminal B
advertisement

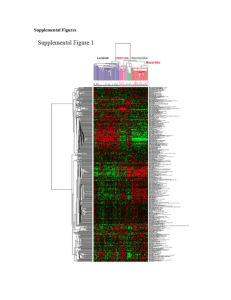
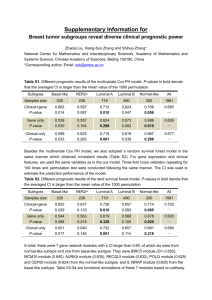
Enabling biomarker validation in breast cancer molecular subtypes: sensitivity and specificity of array-based subtype classification in 983 patients Balázs Győrffy and András Lánczky Research Laboratory for Pediatrics and Nephrology, Hungarian Academy of Sciences - Semmelweis University 1st Dept. of Pediatrics, Budapest, Hungary Objective Abstract Background: The diverse breast cancer molecular subtypes have different clinicopathological characteristics and outcomes. Here, we assessed the correlation between microarray-defined and author-reported molecular subtypes using publicly available breast cancer datasets. Methods: GEO was searched for datasets where the authors determined molecular subtypes. The molecular subtype was re-calculated for each patient using the StGallen guidelines by assessing the microarray-based expression of ER, HER2 and MKI67 (Basal: ER negative + HER2 negative, Luminal A: ER positive + HER2 negative + low MKI67, HER2 enriched: HER2 positive + ER negative, Luminal B: ER positive + HER2 positive and ER positive + HER2 negative + high MKI67). The thresholds used were 500 for ER (probe set 205225_at), 4800 for HER2 (216836_s_at) and 470 for MKI67 (212021_s_at). Sensitivity and specificity were calculated for each subtype separately. Results: Molecular subtype was published in all together six datasets (GSE1456, GSE21653, GSE25066, GSE20711, GSE31519 and GSE17907) for 983 patients. In these, 380, 306, 169 and 128 were Basal, Luminal A, Luminal B and HER2 enriched, respectively. The microarray-based molecular subtype determination resulted in a sensitivity of 0.70, 0.55, 0.63 and 0.38 and a specificity of 0.96, 0.87, 0.67 and 0.99 for Basal, Luminal A, Luminal B and HER2 enriched subtypes, respectively. Finally, the option to filter for molecular subtypes was implemented into our online biomarker validation platform at www.kmplot.com. Discussion: Microarray data provided highest sensitivity and specificity to independent subtype classification for Basal tumors, while the luminal B subtype displayed the highest discordance. Our registration-free online service enables the validation of gene expression based biomarkers in each subtype separately. Background Results What is the correlation between author-reported (IHC-based) and microarray-based molecular subtype determination? GEO search for: studies publishing molecular subtypes raw Affymetrix HGU133A and HGU133plus2 arrays at least 30 patients ER HER2 Luminal A High Low MKI67 N.R. Low Luminal B High High Low High High 1. Highest sensitivity and specificity: Basal tumors GEO datasets: GSE1456, GSE21653, GSE25066, GSE20711, GSE31519 and GSE17907 2. Highest discordance: Luminal B N.R. RFS OS LN+ EndocrineTh. ChemoTh. Basal 3.31 6.00 51% 8.8% (375) 87% (377) Luminal A 4.61 6.67 51% 95% (291) 73% (293) Luminal B 4.35 6.73 58% 33% (147) 74% (147) HER2 3.87 6.49 64% 35% (100) 95% (104) HER2+ Low High Homepages N.R. Breast cancer molecular subtypes: different pathological characteristics different outcomes different treatment Can be identified by: IHC RT-PCR microarray Supported by the OTKA PD 83154 grant. Annual hazard of recurrence and overall survival according to subtypes (Park et al, Breast, 2012): 3. Use the classification to validate survivalassociated genes in each subtype separately at www.kmplot.com/breast: 2. Relation of published subtype distributions: 2. Definition of molecular subtypes using the StGallen guidelines: Basal Low Low 1. Microarray database Clinical characteristics (n=983): Materials & Methods 1. Summary www.kmplot.com/breast 3. Microarray probe sets used for the classification: Gene ER HER2 Probe 205225_at 216836_s_at Cutoff 500 4800 MKI67 212021_s_at 470 4. Statistics: sensitivity [=TP/(TP+FN)] specificity [=TN/(TN+FP)] assess the effect of 22,277 genes on survival in 2,977 breast cancer patients author: as published in GEO by the authors of the datasets computed: distribution for all arrays in the KM-plotter database www.kmplot.com/ovar assess the effect of 22,277 genes on survival in 1,464 ovarian cancer patients 3. Statistics (array vs. IHC, n=983) Basal Luminal A Luminal B HER2 gyorffy@kmplot.com n 380 306 169 128 Sensitivity 0,70 0,55 0,63 0,38 Specificity 0,96 0,87 0,67 0,99 www.recurrenceonline.com compute the ER & HER2 status and the recurrence score for free using microarrays gyer1-6.sote.hu/gyorffy personal homepage Printed by