Bioinformatics Workshop: Gene Set Enrichment Analysis
advertisement
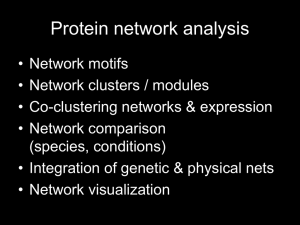
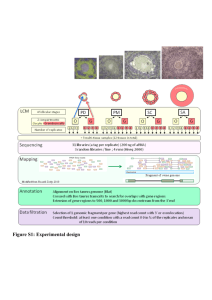
BIOS6660 shRNAseq Gene Set Enrichment Analysis Tzu L Phang PhD Robert Stearman PhD April 16, 2014 Stearman Assessment 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. Genome annotation used was mm9 (from 7/2007). There’s more recent annotation mm10 (12/2011). Was the chr19.fa sequence file derived from mm9 or mm10? It would be nice to show a mapping table that included: Chr19 reads 100% Reads mapped to exons X% Reads not mapping to exons Y% One report I think had the reads mapped to exons numbers but didn’t do anything with it. Of course, what does RNA-seq reads not mapping to exons mean? Four methods used to get overlap gene list. No one had a final table that summarized the values and ranges of fold-change and adjusted p-values. (Some of the FC values were inverse others so this needs to be consistent). No one considered a 3 out of 4 overlap list. No one had a heatmap of the summarized expression values. No one had a supervised cluster based on the overlap genes. Not everyone had R session recorded for reproducible research. Most had a final heatmap of the TOP 15 genes from just the edgeglm method rather than the 14 gene overlap. Several people didn’t get the message about the spliceAlignment argument needed to be on to maximize the reads mapped (~60% vs 90%). If QC reports run and graphs shown but not much in way of interpretation. Some of the heatmap coloring schemes were hard to read and non-standard. Not everyone included a workflow chart to show the analysis path. Tzu Assessment • PDF can not be “knitr” • Try to give more description of what you observe on plot result … Socialism GSEA SuperStar Problems with the SuperStar approach Case 1: No significant genes; because the relevant biological differences are modest relative to the noise inherent to the microarray technology Case 2: Too many significant genes; difficult to interpret and ad hoc approach depends on biologist’s area of expertise Case 3: Single-gene analysis may miss important effects on pathways which normally comprised of sets of genes acting in concert Case 4: Gene lists produced from different labs seldom shown concordances. Gene Set Enrichment Analysis (GSEA) Considers an a priori defined GeneSet (e.g., members of a metabolic pathway), and determines where these members are significantly over-represented or enriched at the top (or bottom) of a list of markers ranked by the degree of correlation with a specific phenotype or class distinction Genes Samples The rows represent the samples or chips, and the columns represent the genes Highly expressed in diseased Diseased Normal Genes on the left side are highly expressed on the top half (indicated by red color) and lowly expressed on the bottom half (indicated by blue color). The reverse is shown on the right-most genes Created a gradient or ranked list corresponding to the degree of correlation with the two phenotypes Lowly expressed in diseased This is depicted nicely by the graph on the bottom of the figure, where the positive ranks on the left represent the correlation to the Disease phenotype and the negative ranks on the right signify the correlation to the Normal phenotype The graph also generates a rank gradient that represents the order of the most up-regulated genes for the Disease sample on the left-most, and the most up-regulated genes for the Normal samples on the rightmost Diseased Normal Now, let’s hide the heatmap and replace the middle part of the figure with genes from a specific geneset, say genes from the Glycolysis pathway. Each vertical blue bars represents a gene from the pathway, being mapped on the same location as the whole dataset Again, genes that are located on the left side are highly expressed on the Disease samples, and the opposite is true for the right-most genes Now, we are ready to demonstrate the GSEA algorithm. The walk down algorithm basically scans the ranked gene list L, and when a member of S is encountered, an Enrichment Score (ES) is registered. This is illustrated on the top part of the figure below; when the ES started to build upon encountering more genes from the GeneSet S. The more S genes is found, the higher the ES But, when no S genes were encountered for a long walk down, as indicated on the middle section of the middle plot, the ES will decrease accordingly. In other words, a high ES relies intimately with the clustering of S genes in close proximity. In this example, we would conclude that the S genes have high degree of correlation with the Disease phenotype since most of the ES was gained from the left portion of the plot Advantages of GSEA • Agnostic to the type of gene set and the source of annotation • Operates on any ordered gene list • Does not require the choice of a gene selection threshold or the explicit definition of a statistically significant marker set • Uses distribution-free, non-parametric, permutation-based test procedures with increased statistical power • Incorporates the permutation of phenotype labels thereby preserving the “biological” correlation structure of the markers • Takes into account multiple hypotheses testing of multiple gene sets References Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S. & Mesirov, J. P. (2005) Proc. Natl. Acad. Sci. USA 102, 15545-15550. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles GSEA Broad Institute (MIT) http://www.broadinstitute.org/gsea/index.jsp GSEA download BioC: gage package • BIOS6660_Share/Week12_13_shRNAseq – Week12_13_shRNAseq_Day2.R – Gage.pdf – We will be using built in dataset. • Direct Download Now, a Demo Mark’s data • BIOS6660_Share/Week12_13_shRNAseq – cep701_AllshRNA_readCounts.txt – Jihye_shRNA_lib_ALL_new.txt cep701_AllshRNA_readCounts.txt Jihye_shRNA_lib_ALL_new.txt Convert Symbol to Entrez