Document
advertisement

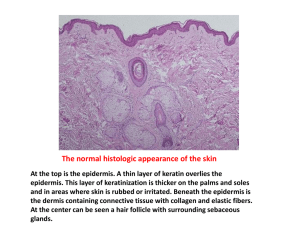
Machine Classification of Melanoma and Nevi from Skin Lesions John David Osborne and Song Gao Outline Melanoma Problem Description Biology Clinical Features Previous Research Methods Sample Images in Experiments Steps in Proposed Algorithm Experimental Results Discussion Limitations of Current Work Future Directions Melanoma and Nevi Melanoma is a form of skin cancer Not the most common, but the most malignant 75% of all skin cancer fatalities Melanocytes (cells that produce the pigment melanin) become cancerous Environmental (sunlight exposure) and genetic influences Nevi or Naevi Singular term is “Nevus” Benign tumor of melanocytes Often confused with melanoma Melanoma versus Nevus Melanoma Nevi Previous Classification Research Extensive research done with detect differences from clinically captured digital images of suspicious moles K-nearest-neighbor classifier was shown to have a mean sensitivity of 95% and 98%, mean specificity of 78% and of 79% on melanoma and nevi respectively. Kernel Logistics PLS classifier was shown to have performance similar to dermatologists with a sensitivity of 95% and a specificity of 60% Another separate recent publication has claimed accuracy over 95% Problem Remains Specificity isn’t that good Consequences are serious Gold standard is histological slide Histology: Melanoma versus Nevi Melanoma Nevus Histological Criteria lesion asymmetry poor circumscription of the lesion with single cells extending beyond the dermal component irregular and confluent nests variable in size and shape pagetoid spread (suprabasal melanocytes) confluent growth and the dermal epidermal junction (DEJ) absence of maturation (failure of melanocyte nuclei to become smaller with further descent into the nucleus) involvement of the hair follicle cytological and nuclei atypia nuclear enlargement melanocytic nuclei enlarged relative to keratinocytic ones variability in size and shape of nucleus hyperchromatism and prominent nucleoli) mitoses in the dermis and the presence of dermal necrotic melanocytes [1, 2]. Previous Work Histology Image Processing Minimal Much more complicated Neuroblastoma Intracellular protein localization Determination of malignancy based on staining pattern Automated identification of abnormal metaphase chromosome cells for the detection of chronic myeloid leukemia using microscopic images To the best of our knowledge, no other group is distinguishing between melanoma and nevus on the basis of histology slides Sample Images in Experiments 40x 100x 200x 400x Components in Case Image Steps in Proposed Algorithm Remove irrelevant areas 1. Coarse filtering Fine filtering – more accurate! Recovering Distinguish relevant areas 3. SVM training for the prediction 2. Extract features SVM model training Coarse Filtering All images are converted into HSV color space, which is more perceptual uniform compared with RGB color space. 120° 0 0 ° red tissue Slide area 240° nucleus Thresholdh=0.7, (252°) Thresholds=0.05 6th degree 10th degree Fine Filtering Red area occupies most remaining pixels of the image after coarse filtering. A precise threshold is required to further remove the red area. A histogram with 256 bins is built based on the h-value of the remaining pixels. The interval of dominant bin represents the h-range of red tissue. Pixels with h-value above this interval are all related with red tissue. Polynomial curve fitting – p(x) Better describe the distribution of the histogram The higher the degree is, the better the curve fits. argmax(p(x)) is more accurate as a threshold than the interval value of the highest bin. Pixels with h-value larger than argmax(p(x)) is removed from the image. Recovering (1/2) The nuclear stain bleeds over into the surrounding giving them a similar hue and saturation to the red tissue. The surrounding pixels with the form of small segments are also removed by the filtering procedure. Small segments are related with cytoplasm, which is relevant area. Need recover the small segments Mask after filtering White: irrelevant area Recovering (2/2) What’s the area threshold? A histogram is built based on the descending area of segments. Bin width – area interval (e.g. 10 pixels) Bin height – # of segments within corresponding area interval The area threshold is determined by the area interval of the 1st bin which has a lower number of segments than an user input parameter. (e.g. 10 segments) Demo of Image Segmentation (a) Original Image | # of pixels: 3133440 (c) (d) (b) After Filtering # of pixels: 2002070 After Color Recovering | #| of pixels: 1950855 After Coarse Filtering | # of pixels: 1740271 Coarse filtering Fine filtering (e) Recovering Otsu’s method Distinguish between blue area and white area Relevant areas Extracting Features Criteria The size of a nucleus becomes larger within melanocytes. 2. The shape of nucleus of a melanocyte tends to become more asymmetric. 1. Four features The ratio of the number of nuclei to the area of cytoplasm 2. The ratio of the area of nuclei to the area of cytoplasm 3. The ratio of the perimeter of a nucleus to its area 4. The ratio of the major length of a nucleus to its minor length 1. Minor axis Major axis Major axis The SVM Training for the Prediction A multi-class support vector classification (SVC) is provided by LIBSVM* Two kernel functions are used in the SVC, such as rbf (Radial basis function) function and linear function. 15 feature combinations with 4 different magnification of training dataset are trained on SVC. *http://www.csie.ntu.edu.tw/~cjlin/libsvm/ Experiments (1/2) Statistical information of image dataset Magnification 40x 100x 200x 400x total # of images 31 33 32 30 126 # of melanoma 18 18 18 18 72 # of nevus 13 15 14 12 54 Evaluation criteria Accuracy: # of correctly predicated records Melanoma – positive; nevus – negative specificity # of True Negatives # of True Negatives # of FalsePositives sensitivity # of True Positives # of True Positives # of False Negatives Experiments (2/2) SVC comparative results SVC model with diff kernel and feature combinations 400x Accuracy Specificity Sensitivity nu-SVC rbf [3+4] 0.90 0.75 1.0 nu-SVC rbf [2+3+4] 0.87 0.67 1.0 nu-SVC rbf [1+2+3+4] 0.87 0.67 1.0 nu-SVC linear [3+4] 0.87 0.83 0.89 nu-SVC linear [2+3+4] 0.87 0.83 0.89 nu-SVC linear [1+2+3+4] 0.90 0.83 0.94 Limitations of Current Features Area Features The number of nuclei to the area of cytoplasm. Each segment in the blue area is a nucleus, and the area of the white area is the area of cytoplasm. The area of nuclei to the area of cytoplasm Atypical Nucleus Features The perimeter of a nucleus to the area of it. The major length of a nucleus to the minor length of it Our feature selection has 2 major problems Melanocyte Problem Lymphocyte Problem The Melanocyte Problem -Relies on someone else to fill the image with melanocytes -Not really automated -Won’t work on all areas of the slide -Nuclear atypia in nonmelanocytes has nothing to do with melanonoma 2 2 The Lymphocyte Problem Inflammatory infiltrate is full of lymphocytes with staining nucleus Lymphocytes are very small, and a dense distribution of nuclei Indicates inflammation, present in various disease and infections including melanoma Looking for nuclear superploidy, may be counting lymphocytes 2 3 Limitations of Current Work Using only a tiny fraction of available known features Over a dozen features available Lack of feedback for performance metrics for cell type identification and ground truth Are we looking at the right cells? Have melanocyte masks, could use them Lack of regional information Is this melanocyte in the right place Reliance on human to set slide Inability to classify the various types of melanoma or nevi Melanocyte Taxonomy Nevus Variants Melanoma Variants Becker’s Nevus Nevoid Melanoma Junctional Nevus Small cell melanoma Compound Nevus Desmoplastic melanoma Banal Nevus (has numerous variants) Melignant blue nevus Dysplastic Nevus Pigment-synthesizing melanoma Balloon cell nevus Rhabdoid melanoma Meyerson’s nevus Myxoid melanoma Halo (Sutton’s) nevus Adenoid (psuedoglandular) melanoma Recurrent nevus (psuedomelanoma) Angiotripic (pseudovascular) melanoma Inverted type A (clonal) nevus Signet-ring cell melanoma Cockarde nevus Balloon cell melanoma Nevus spilus Clear-cell melanoma Collision tumors Metaplastic Melanoma Eccrine-centered nevus Spitzoid melanoma Acral nevus Giant cell melanoma Future Directions Current work Used simple (banal) melanoma and some cases of melanoma in situ Simple banal nevus and one dysplastic nevus Slides were manually processed to select areas of interest Obvious extensions Handle more types of Melanoma and Nevi Additional diagnostic features Get and use the entire slide Ability to recognize cell types Ability to recognize regions and layers Maturation of Nests Additional Features A - Asymmetry in an intradermal naevus (25x) B – Lymphocyte Infiltration (100x) C - Confluent nests in a junctional nevus (200x) D - Poor circumscription: the junctional melanocytic proliferation ends with single cells (200x) E - Predominance of single melanocytes and suprabasal melanocytes (200x) F - Involvement of the hair follicle G - Cytological atypia in a compound naevus (300x) 2 7 Lowest Hanging Fruit Additional Feature - Lesion Asymmetry Highly diagnostic Need entire slide Cell type recognition Melanocyte Lymphocyte (small, darkly staining) Can use additional feature of lymphocyte invasion Keratinocytes Epithelial cells Region detection Allows detection of nesting, maturation and pagetoid spread Nevus Melanoma Highest Hanging Fruit Multiple types of nevus and melanoma Need knowledge base of diagnostic criteria Could make the algorithm more general purpose (allow detection of melanoma mimics) 3 0