Introduction to Mathematica
advertisement

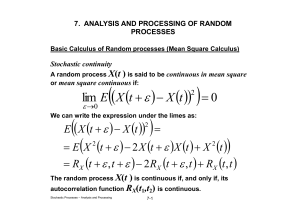
Simulation of Biochemical Reactions for Modeling of Cell DNA Repair Systems Dr. Moustafa Mohamed Salama Laboratory of Radiation Biology, JINR Supervisor : Dr. Oleg Belov Simulation of Biochemical Reactions Stochastic approach Master Equation Deterministic Approach Exact Stochastic Simulation Reaction-Based Solving Methods: • We are used to writing differential equations from chemical reactions. Is converted to • For example: X+Y Z (rate a) ZY (rate b) dX/dt = -aXY; dY/dt = -aXY +bZ; dZ/dt = aXY-bZ; • But in stochastic systems the actual “events” or “reactions” is stochastic. • And, when a reaction occurs, it affects many “chemicals” at once. 3 Stochastic? • “Random or Probabilistic“ • Stochastic simulation: uses a random number generator to produce one or more possible time courses. Monte Carlo Simulations: Stochastic Simulation Algorithm General Form of Algorithm Entire Simulation Input cʋ (ʋ=1,…,M) initi . Of Xi (i=1,…,N) Set t=0 & n=0 Generate random numbers r1 and r2 Calculate a1= hvcʋ (ʋ=1,…,M) a0 = aʋ Generate random numbers r1 and r2 t Take 1 1 • • • 1 a0 1 ln r 1 a i r2 a 0 a i 1 Update t = t + t Update X = [X1, X2, …XC] Update n= n + 1 OR Stop If t > tstop no more Reactants Remain (hv =0) Step 1: Given the system state, determine the rate of each reaction, aʋ . • Reaction 1: S1 + S2 S3, with rate constant c1 – X1, X2 are the numbers of the reactant molecules – Define the stoichiometry: h1 = X1X2 ; this will give dependence on amounts of molecules. – Then a1= h1c1= k1 X1X2 = rate for this reaction. • Reaction 2: S1 + S1 S2, – h2 = X1(X1-1)/2 • Finally, define: a0 = aʋ (ʋ = 1 to M) – This is the combined rate of all possible reactions 7 Step 2 When does the next reaction occur … r1 8 1 0.8 0.6 0.4 0.2 • This is time of the next event. • (Note that the time step doesn’t have to be predetermined, and is exact.) t • Let 1 t ln a 0 r1 1 16 14 12 10 8 6 4 2 0 0 • Pick r1, a uniform random number from 0 to 1 Step 2 …and which reaction is it? • Determine which reaction occurs at time t: • Pick r2, another uniform random number from 0 to 1 1 • Find , such that: a 1 i r2 a 0 a i 1 • Think about dividing a0 into M pieces of length aʋ 9 Step 3 Update the System State Step 3 is to determine how each of C chemicals are affected • Update t = t + t • Update X = [X1, X2, …XC] according to the reaction stoichiometry • Update reaction step counter. • If t > tstop or if no more reactions remain ( all (hv =0)), terminate the calculations ; otherwise, return to step1. 10 Why consider Mathematica? • Powerful system for symbolic mathematical but also handles numerical mathematics, graphics, data visualization, simulation. • Larger community of users comparing with others. • Containing the toolkits of Stochastic Simulation Algorithm (SSA) Example in Mathematica Example in Mathematica Example in Mathematica Mathematical modeling of repair of DNA Single strand breaks in Escherichia coli bacterial cells By: Mohamed Abd Elmoez Type I Repair Complex between un legated DNA and Ligase DNA Ligase Repaired DNA Mathematical modeling of recombination repair mechanism for Double strand DNA breaks in Escherichia coli bacterial cells by : Alla Mohamed RecBCD complex concentration change N N t t Conclusion and Future work • We learned here how to make a Mathematical modeling for the chemical reactions. • Know more features about Tools in Mathematica software toolkits of Stochastic Simulation Algorithm. • We discussed developing a new algorithm for Stochastic approach for range in rate of reactions. Acknowledgment •I ‘d like to thank JINR especially Summer school members. •I also wish to thank Dr. Belov for Fruitful discussions on Mathematical modeling in radiation biology.