Slides 1
advertisement

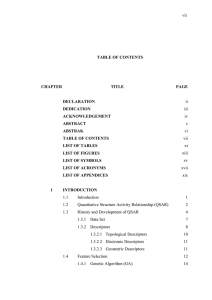
Quantitative Structure-Activity Relationships Quantitative Structure-Property-Relationships SAR/QSAR/QSPR modeling Alexandre Varnek Faculté de Chimie, ULP, Strasbourg, FRANCE SAR/QSAR/QSPR models • Development • Validation • Application Classification and Regression models • Development • Validation • Application Development of the models • • • Selection and curation of experimental data Preparation of training and test sets (optionaly) Selection of an initial set of descriptors and their normalisation Variables selection (optionally) Selection of a machine-learning method • • Validation of models • • Training/test set Cross-validation - internal, external Application of the Models • Models Applicability Domain Development the models • • • • Experimental Data: selection and cleaning Descriptors Mathematical techniques Statistical criteria Data selection: Congenericity problem • Congenericity principle is the assumption that « similar compounds give similar responses ». This was the basic requirement of QSAR. This concerns structurally homogeneous data sets. • Nowdays, experimentalists mostly produce structurally diverse (non-congeneric) data sets Data cleaning: • • • • • Similar experimental conditions Dublicates Structures standardization Removal of mixtures ….. The importance of Chemical Data Curation Dataset curation is crucial for any cheminformatics analysis (QSAR modeling, clustering, similarity search, etc.). Currently, it is uncommon to describe procedures used for curation in research papers; procedures are implemented or employed differently in different groups. We wish to emphasize the need to create and popularize standardized curation strategy, applicable for any ensemble of compounds. What about these structures? (real examples) Why duplicates are unsafe for QSAR ? Duplicates are identical compounds present in a given dataset. OH CH3 HO CH3 CH3 CH3 OH H3C H3C CH3 OH CH3 OH CH3 OH ID = 256 ID = 879 ID = 2346 Manual identification of duplicates is practically impossible especially when the dataset is large. Activity analysis of duplicates is also highly important to identify cases where one occurrence is identified as ‘active’ and another one as ‘weak active’ or ‘inactive’. CH3 CH3 HO OH H3C ACTIVE CH3 INACTIVE H3C OH OH CH3 Structural standardization For a given dataset, chemical groups have to be written in a standardized way, taking into account critical properties (like pH) of the modeled system. Aromatic compounds OH OH These two different representations of the same compound will lead to different descriptors, especially with certain fingerprint or fragmental approaches. Cl Cl CH3 CH3 Carboxylic acids, nitro groups etc. O O HO X O– O O O X X O O OH N N+ X X For a given dataset, these functional groups have to be written in a consistent way to avoid different descriptor values for the same chemical group. Normalization of carboxylic, nitro groups, etc. removal of inorganics All inorganic compounds must be removed since our QSAR modeling strategy includes the calculation of molecular descriptors for organic compounds only. This is an obvious limitation of the approach. However the total fraction of inorganics in most available datasets is relatively small. To detect inorganics, several solutions are available: - Automatic identification using in combination Jchem (ChemAxon, cxcalc program) to output the empirical formula of all compounds and simple scripts to remove compounds with no carbon; - Manual inspection of compounds possessing no carbon atom using Notepad++ tools. removal of mixtures Fragments can be removed according to the number of constitutive atoms or the molecular weight. removal of mixtures However, some cases are particularly difficult to treat. Examples from DILI - BIOWISDOM dataset: ID=172 CLEANED FORM BY CHEMAXON The two eliminated compounds could be active ! . INITIAL FORM MANUAL INSPECTION/VALIDATION IS STILL CRUCIAL ID=1700 INITIAL FORM CLEANED FORM BY CHEMAXON Ok. removal of salts Options Remove Fragments, Neutralize and Transform of Chemaxon Standardizer. have to be used simultaneously for best results. Aromatization and 2D cleaning ChemAxon Standardizer offers two ways to aromatize benzene rings, both of them based on Hűckel’s rules. “General Style” O CH3 NH OH “Basic Style” CH3 O NH OH Most descriptor calculation packages recognize the “basic style” only. http://www.chemaxon.com/jchem/marvin/help/sci/aromatization-doc.html Preparation of training and test sets Building of structure property models Training set Initial data set Test 10 – 15 % Splitting of an initial data set into training and test sets Selection of the best models according to statistical criteria “Prediction” calculations using the best structure property models Recommendations to prepare a test set • (i) experimental methods for determination of activities in the training and test sets should be similar; • (ii) the activity values should span several orders of magnitude, but should not exceed activity values in the training set by more than 10%; • (iii) the balance between active and inactive compounds should be respected for uniform sampling of the data. References: Oprea, T. I.; Waller, C. L.; Marshall, G. R. J. Med. Chem. 1994, 37, 2206-2215 Descriptors • Variables selction • Normalization molecules descriptors Pattern matrix Selection of descriptors for QSAR model QSAR models should be reduced to a set of descriptors which is as information rich but as small as possible. Objective selection (independent variable only) Statistical criteria of correlations Pairwise selection (Forward or Backward Stepwise selection) Principal Component Analysis Partial Least Square analysis Genetic Algorithm ………………. Subjective selection Descriptors selection based on mechanistic studies Preprocessing strategy for the derivation of models for use in structure-activity relationships (QSARs) 1. identify a subset of columns (variables) with significant correlation to the response; 2. remove columns (variables) with zero (small) variance; 3. remove columns (variables) with no unique information; 4. identify a subset of variables on which to construct a model; 5. address the problem of chance correlation. D. C. Whitley, M. G. Ford, D. J. Livingstone J. Chem. Inf. Comput. Sci. 2000, 40, 1160-1168 Descriptors Normalisation descriptors n molecules m j (1/ n) xij* i 1 n s (1/ n) ( xij* m j ) 2 2 j i 1 Pattern matrix Normalisation 1 (Unit Variance scaling): Normalisation 2 (Mean Centring Scaling): xij x m j * ij xij xij* m j sj Data Normalisation Initial descriptors Norm. 1 Norm. 2 Machine-Learning Methods Fitting models’ parameters Y = F(ai , Xi ) Xi - descriptors (independent variables) ai - fitted parameters The goal is to minimize Residual Sum of Squared (RSS) N RSS ( yexp,i ycalc,i ) i 1 2 Multiple Linear Regression Activity Descriptor Y1 X1 Y2 Y2 … … Yn Xn Yi = a0 + a1 Xi1 Y X Multiple Linear Regression y=ax+b Residual Sum of Squared (RSS) N RSS ( yi ycalc,i ) i 1 2 b a Multiple Linear Regression Activity Descr 1 Descr 2 … Descr m Y1 X11 X12 … X1m Y2 X21 X22 … X2m … … … … … Yn Xn1 Xn2 … Xnm Yi = a0 + a1 Xi1 + a2 Xi2 +…+ am Xim kNN (k Nearest Neighbors) Activity Y assessment calculating a weighted mean of the activities Yi of its k nearest neighbors in the chemical space TRAINING SET Descriptor 1 Descriptor 2 A.Tropsha, A.Golbraikh, 2003 Biological and Artificial Neuron Multilayer Neural Network Neurons in the input layer correspond to descriptors, neurons in the output layer – to properties being predicted, neurons in the hidden layer – to nonlinear latent variables SVM: Support Vector Machine w, x b 1 w, x b 0 w, x b 1 2 w Support Vector Classification (SVC) SVM: Margins The margin is the minimal distance of any training point to the separating hyperplane Margin 1 w Support Vector Regression ε-Insensitive Loss Function Only the points outside the εtube are penalized in a linear fashion 0 if : otherwise Kernel Trick In low-dimensional input space K ( x, x) ( x), ( x) In high-dimensional feature space Any non-linear problem (classification, regression) in the original input space can be converted into linear by making non-linear mapping Φ into a feature space with higher dimension QSAR/QSPR models • Development • Validation • Application Preparation of training and test sets Building of structure property models Training set Initial data set Test 10 – 15 % Splitting of an initial data set into training and test sets Selection of the best models according to statistical criteria “Prediction” calculations using the best structure property models Validation Estimation of the models predictive performance 5- Fold Cross Validation All compounds of the dataset are predicted Dataset Fold1 Fold2 Fold3 Fold4 Fold5 Leave-One Out Cross-Validation N- Fold Internal Cross Validation • Cross-validation is performed AFTER variables selection on the entire dataset. • On each fold, the “test” set contains only 1 molecule Statistical parameters for Regression 42 Fitting vs validation Stabilities (logK) of Sr2+L complexes in water LogKcalc LogKpred 12 LOO 12 Fit 9 6 0 9 9 6 6 3 R2 = 0.886 RMSE = 0.97 3 12 3 R2= 0.826 RMSE = 1.20 0 5-CV R2 = 0.682 RMSE = 1.62 0 -3 0 3 6 9 12 15 0 3 6 9 12 15 3 6 9 12 15 LogKexp All molecules were used for the model preparation Each molecule was “predicted” in internal CV Each molecule was predicted in external CV Regression Error Characteristic (REC) REC curves are widely used to compare of the performance of different models. The gray line corresponds to average value model (AM). For a given model, the area between AM and corresponding calculated curve reflects its quality. Statistical parameters for Classification Confusion Matrix Classification Evaluation sensitivity = true positive rate (TPR) = hit rate = recall TPR = TP / P = TP / (TP + FN) false positive rate (FPR) FPR = FP / N = FP / (FP + TN) specificity (SPC) = True Negative Rate SPC = TN / N = TN / (FP + TN) = 1 − FPR positive predictive value (PPV) = precision PPV = TP / (TP + FP) negative predictive value (NPV) NPV = TN / (TN + FN) accuracy (ACC) ACC = (TP + TN) / (P + N) balanced accuracy (BAC) BAC = (sensitivity + sensitivity ) / 2 = (TP / (TP + FN) + TN / (FP + TN)) /2 Receiver Operating Characteristic (ROC) TPR Plot of the sensitivity vs (1 − specificity) for a binary classifier system as its discrimination threshold is varied. The ROC can also be represented equivalently by plotting the fraction of true positives (TPR = true positive rate) vs the fraction of false positives (FPR = false positive rate). FPR Ideally, Area Under Curve (AUC) => 1 ROC (Receiver Operating Characteristics) 100% TP 0 FP a 1 2 3 b c d 4 5 6 7 8 9 e f g h i j FN 0 TN a 1 2 3 b c d 4 5 6 7 8 9 e f g h i j TP% Ideal model: AUC=0.84 AUC=1.00 j g Useless model: AUC=0.50 0% FP% 0 a 1 b h 100% 2 3 d 6 e f i 5 c 8 4 7 9 When a model is accepted ? Regression Models Classification Models 3 classes Determination coefficient R2 > R02 Here, R02 = 0.5 49 BA > 1/q for q classes “Chance correlation” problem 2,000 1 1,500 0.75 1,000 0.5 1965 1970 1975 year 1980 a model MUST be validated on new independent data to avoid a chance correlation Y-Scrambling (for methods without descriptor selection) X1 X2 Y1 Y2 Y2 Y5 X3 Y3 Y4 X4 Y4 Y6 X5 Y5 Y1 X6 Y6 Y7 X7 Y7 Y3 R2 0.0 1.0 Y-Scrambling (for methods without descriptor selection) X1 X2 Y1 Y2 Y4 Y1 X3 Y3 Y5 X4 Y4 Y2 X5 Y5 Y6 X6 Y6 Y3 X7 Y7 Y7 R2 0.0 1.0 Y-Scrambling (for methods without descriptor selection) X1 X2 Y1 Y2 Y7 Y6 X3 Y3 Y3 X4 Y4 Y5 X5 Y5 Y4 X6 Y6 Y1 X7 Y7 Y2 R2 0.0 1.0 QSAR/QSPR models • Development • Validation • Application Test compound QSPR Models Prediction Performance Robustness of QSPR models - Descriptors type; - Descriptors selection; - Machine-learning methods; - Validation of models. Applicability domain of models Is a test compound similar to the training set compounds? Applicability domain of QSAR models Descriptor 2 The new compound will be predicted by the model, only if : Di ≤ <Dk> + Z × sk with Z, an empirical parameter (0.5 by default) TRAINING SET Descriptor 1 = TEST INSIDE THE DOMAIN OUTSIDE THE DOMAIN Will be predicted Will not be predicted COMPOUND Applicability domain of QSAR models Applicability Domain Approaches Fragment –based methods Fragment Control (FC) Density based methods 1-SVM Model’s Fragment Control (MFC) Distance –based methods zkNN Range –based methods Bounding Box (BB) Ensemble modeling Hunting season … Single hunter Hunting season … Many hunters Ensemble modelling Ensemple modeling Y1 Y2 1 n Consensus = Y i n i 1 Y3 Screening and hits selection Database O COOH Cl Br OH N OH Virtual Sreening N OH QSPR model N COOH Useless compounds O Br Hits Experimental Tests