(31ºN), in the
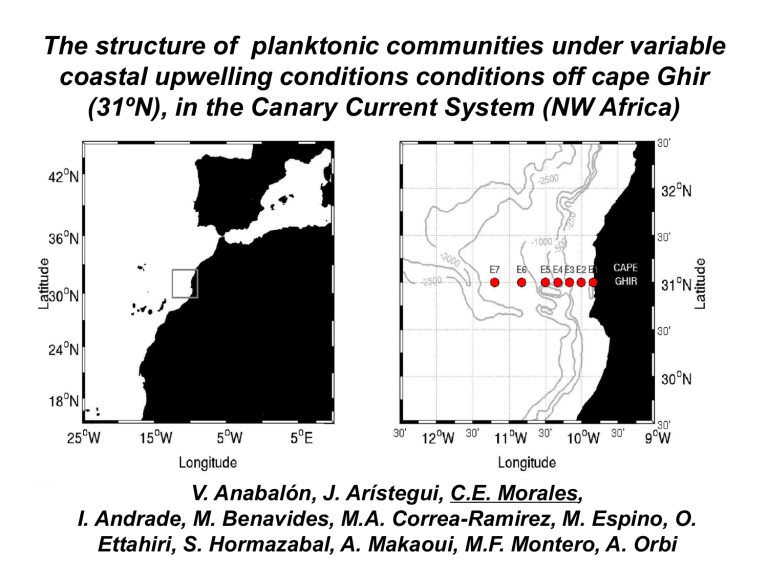
The structure of planktonic communities under variable coastal upwelling conditions conditions off cape Ghir
(31ºN), in the Canary Current System (NW Africa)
V. Anabalón, J. Arístegui, C.E. Morales,
I. Andrade, M. Benavides, M.A. Correa-Ramirez, M. Espino, O.
Ettahiri, S. Hormazabal, A. Makaoui, M.F. Montero, A. Orbi
Background
Area of permanent upwelling, narrow shelf, fronts & filaments, low NO3 concentration compared to other areas and regions.
Pelegri et al. 2005
MOTIVATION
Do changes in upwelling intensity produce significant spatiotemporal variations in the structure of planktonic communities
(coastal and coastal transition zones -CTZ)?
Alonshore wind stress
SST
Chl-a
Samplings
APPROACHES AND METHODS
Oceanographic cruises (5): Dec-2008; Feb-, June, Aug,
Oct-2009; transect perpendicular to the coast (7 stations, coast to app. 150 Km offshore.
- Hydrographic data: CTD with fluorescence sensor. Estimates of water density (as sigma-t) and stratification intensity (J m-3) according to Bowden (1983).
- Seawater samples at 5 levels (0, 25, maximum fluorescence depth, 90, 150 m depth):
Niskin bottles (5 L); analyses:
* Macro-nutrients (NO2+NO3, PO4, Si)
* Chla (total, <20 and <3 µm)
* Micro-organisms: picoplankton (flow-cytometry; only 3 cruises), nanoplankton
(epifluorescence and
Utermöhl), and microplankton (Utermöhl).
Satellite time series data: the wider perspective
- Winds
(CCMP; ¼ ° x ¼ ° resolution);
- SST (AVHRR Pathfinder V5.0 from NOAA; ftp://data.nodc.noaa.gov/pub/data.nodc/Pathfinder) at 2x2 Km resolution;
- Sea level anomaly (combined processing of TOPEX/JASON at ¼ ° x ¼ ° resolution
- ERS altimeter data distributed by AVISO (http://aviso.oceanos.com)
surface geostrophic flow field;
- Chl-a form HERMES (combined sensors (MODIS, MERIS, SeaWiFS), obtained from GlobColorWeb (ftp.fr-acri.com).
APPROACHES AND METHODS (2)
Plankton biomass (C):
- Nanoplankton and microplankton: geometric models for cell volume estimates
(Chrzanowski & Simek, 1990; Sun & Liu 2003). C/biovolume conversion factors:
Menden-Deuer & Lessard (2000) for CIL, DIN, and DIAT; Heinbokel (1978) for
Tintinnids; and Borsheim & Bratbak (1987) for FLA.
- Autotrophic picoplankton: 29 fg C/cell - PRO, 100 fg C/cell - SYN (Zubkov et al., 2000),
1.5 pg C/cell - PEUK (Zubkov et al., 1998); HB: 12 fg C/cell (Fukuda et al., 1998).
- Mixotrophy (DIN + CIL), literature recognition of mixotrophy at species/genus level
(40% autotrophy) in the case of microplankton (no autofluorescence data available).
Statistics: multivariate analyses, PRIME software v.6
(Clarke & Warwick, 2001; Clarke & Gorley, 2006)
- MDS (nonmetric multidimensional scaling) for cluster identification; hydrographic and biological matrices; significance of the clusters
– SIMPROF
- ANOSIM for analysis of similarities; SIMPER for groups/species contributions to similarities and dissimilarities between clusters in the biological matrix.
- BIO-ENV and RELATE to analyze the associations between the biological data and the environmental variables. The best combinations of variables determined by BIO-ENV were subjected to further analysis (LINKTREE) to identify the variable(s) which best represented the separation of the biological components into different groups/cluster.
WEUP
SST: 1617ºC
SST grad.: <2ºC
Wind: 8-12 m/s NE
Low stability
RELAX
SST: 18ºC
SST grad.: 3.5ºC
Wind: 4-8 m/s NW
MOUP
SST: 1920ºC
SST grad.: 4ºC
Wind: 4-8 m/s NE
Shoaling of isopycnals at the coast & counterflow
HYDROGRAPHIC CLUSTERS
Variables contribution to cluster separation:
- nutrient concentration: WEUP vs. E1
- nutrient concentration and SST: MOUP vs. E1
- water density , SST and Nº days favourable to upwelling:
WEUP vs. MOUP
Cross-shore variability (E2-E4 vs. CTZ E5-E7 stations)
BIOLOGICAL CLUSTERS
DINOFLAGELLATES (43%) contributed most to cluster separation (E1 vs. rest).
DIATOMS (21%) and
CILIATES (21%).
Dissimilarity between:
WEUP - RELAX:
DINOFLAGELLATES (38%) and CILIATES (32%).
WEUP - MOUP:
DINOFLAGELLATES (37%) and DIATOMS (34%).
RELAX - MOUP: DIATOMS
(34%); CILIATES (24%) and
DINOFLAGELLATES (22%).
• Inclusion of the picoplankton fraction (only 3 samplings): minimal influence in terms of biomass.
BIOMASS: micro+nanoplankton
Dominance of microplankton
(>53%): DINOFLAGELLATES
+ CILIATES
Nanoplankton:
DINOFLAGELLATES +
FLAGELLATES
Autotrophic-C: DIATOMS exceptions
Dec-08: APP, AFL, ADIN
Aug-09: ADIN + DIAT
As Chl-a: nanoautotrophs.
Heterotrophic-C: DINOFLAG.
RELEVANCE OF MIXOTROPHY
Mean H:A biomass ratios (pico-to micro): 3 samplings
- No correction mixotrophy: >1 (inverted pyramid)
- Correction for mixotrophy: <1 (normal pyramid)
BIOMASS-C STRUCTURE IN CAPE GHIR:
BIOMASS DIFFERENCES (Linktree) micro+nanoplankton
CONCLUSIONS
RESTRICTIONS: SNAPSHOT OF THE SYSTEM
Two main upwelling phases weak (no gradients acrosshore) and moderate (strong crosshore gradients).
Separation of the most coastal station (depth effect). Cluster formation influenced by nutrient concentration (spatial), SST and upwelling constancy (temporal).
Hydrographic clusters were representative of the spatiotemporal variability in planktonic assemblages changes in the upwelling intensity do influence community structure.
The dominant functional groups (C-biomass) were mixed assemblages of DIN and CIL (>51%); DIAT contributions were moderate to low (<35%). Total Chl-a was dominated by the nanoplankton and mixotrophy is important in H:A evaluation for this system.
QUESTIONS UNSOLVED
Is upwelling intensity in the region (NW Africa) increasing or decreasing ???
Is the presence of mixed autotrophic assemblages a consequence of recent changes in upwelling intensity in this region ???
How well can be represent mixotrophs in primary production
(PP) estimates and in ecosystem models??? How well can be represent other types of PP or nutrient requirements ???
Heterotrophic:autotrophic ratios – how well can be estimate the biomass of the diverse components ??? (basic!)
Biomass estimates: Chl-a versus Carbon; relevance in remote sensing estimates of primary production (changing
C:Chl-a ratios)
Sampling A/ Chl-a total Am/ Chl-a total MAT/Chl-a micro MAT/ Chl-a micro
Dec-08 C1
Feb-09 C2
Jun-09 C3
Aug-09 C4
Oct-09 C5
50
50
64
60
60
80
30
100
310
330
90
74
140
390
450
160
NAT/Chl-a nano APP/ Chl-a pico
31
75
42
20
14
148
162
27
All samplings A/Chl-a total Am/ Chl-a total MAT/ Chl-a
REGRESSIONS
Micro MATm/ Chl-a Micro NAT/ Chl-a Nano APP/ Chl-a Pico y = 34.8x +926
R² = 0.49 p= 0.001
y = 44.0x+1122
R² = 0.58 p= 0.0007
y = 61.3x + 528
R² = 0.35
p = 0.1
y = 92.5x + 1127
R² = 0.51
p = 0.05
y = 44.9x -172
R² = 0.60 p=0.0001
y = 89.9x + 311
R² = 0.59 p = 0.0001