DNA barcoding the flora of Wales
advertisement
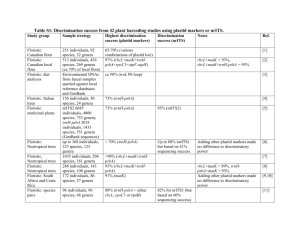
Barcode Wales: DNA barcoding the flora of Wales Natasha de Vere, Tim Rich, Col Ford, Sarah Trinder, Charlie Long, Chris Moore, Danielle Satterthwaite, Helena Davies, Joe Moughan, Addie Griffith, Laura Jones, Joel Allainguillaume, Mike Wilkinson, Kevin Walker, Tatiana Tatarinova, Hannah Garbett, Les Baillie, Jenny Hawkins DNA barcoding • DNA based species identification • Once reference barcodes are established will always be able to identify that species • Internationally agreed regions of DNA used: rbcL & matK for plants • Open science >Cotoneaster_cambricus AGAGACTAAAGCAAGTGTTGGATTCAAAGCTGGTGTTAAAGATT ATAAATTGACTTATTATACTCCTGACTATGAAACCAAAGATACTG ATATTTTGGCAGCATTTCGAGTAACTCCTCAACCTGGAGTTCCAC CTGAGGAAGCAGGGGCCGCGGTAGCTGCTGAATCTTCTACTGGT ACATGGACAACTGTATGGACTGACGGTCTTACCAGTCTTGATCGT TACAAAGGTCGATGCTACCACATCGAGCCTGTTGCTGGAGAAGA AAGTCAATTTATTGCTTATGTAGCTTACCCCTTAGACCTTTTTGAA GAAGGTTCTGTTACTAACATGTTTACTTCCATTGTAGGTAATGTGT TTGGGTTCAAGGCCCTGCGCGCTCTACGTCTGGAGGATTTGCGA ATCCCTACTGCTTATGTTAAAACTTTCCAGGGCCCGCCTCATGGT ATCCAAGTTGAGAGAGATAAATTGAACAAGTATGGCCGCCCTCT ATTGGGATGTACTATAAAACCAAAATTGGGGTTATCCGCTAAGAA TTACGGTAGAGCAGTTTATGAATGTC What do they eat? What’s in this? What pollen is it carrying? Is this legal? Barcode Wales: Cod Bar Cymru • DNA barcode the native flowering plants and conifers of Wales • Develop applications that utilise this research platform Sample collection • 1143 native flowering plants and conifers • 4272 individuals sampled, 3637 herbarium, 635 freshly collected • All specimens verified by taxonomic expert • Herbarium vouchers and full collection details for all samples DNA barcoding protocol • DNA extraction: Qiagen kits, modified for herbarium material • Amplification: rbcL 5 & matK 23 primer combinations • Sanger sequencing • Manual editing, multiple alignment • Sequences uploaded onto BOLD & GenBank – publically available de Vere N, Rich TCG, Ford CR, Trinder SA, Long C, et al. (2012) DNA Barcoding the Native Flowering Plants and Conifers of Wales. PLoS ONE 7(6): e37945. Recoverability rbcL matK rbcL & matK No. of spp. sequenced (1143) 1117 (98%) 1031 (90%) 1025 (90%) No. of spp. with > 1 individual sequenced 1041 (91%) 814 (71%) 808 (71%) Mean no. of individuals per spp. 3 2 2 Mode of individuals per spp. 3 3 3 Range of individuals per spp. 1-9 1-8 1-8 Total no. of individuals sequenced 3304 2419 2349 In total 5,723 barcode sequences obtained for the 1143 species Effect of herbarium specimen age Spearman Rank Correlation: rbcL rho = 0.993*** matK rho = 0.986*** Relative discrimination 808 species with multiple individuals sequenced for both rbcL & matK 69 99 75 68 74 56 57 100 98 99 96 95 rbcL & matK discrimination Scale n Mean discrimination % (SD) 10x10 km 253 82 (3) 2x2 km 1116 93 (6) Species lists generated for each square, discrimination assessed by presence of a barcode gap Roots, shoots and pollen • Root analysis • Identification at single pollen grain resolution • Identification of pollen products Gives a feeling “of Youthful Exuberance!” Honey and drug discovery • Collect honey from throughout the UK. • Test antibacterial properties of honey against MRSA and Clostridium difficile. • DNA barcode honey using next generation DNA sequencing (454). • Identify plant derived phytochemicals that increase antimicrobial activity. MRSA – prelim results DNA barcoding and phylogenetics ML tree for rbcL, RAxML (GTR+CAT) 1000 bootstraps, on the CIPRES supercomputer cluster 56% of species form monophyletic groups 44% with bootstrap support >70% DNA barcoding and phylogenetic ecology ML tree for rbcL, threatened species traced using Mesquite What’s next? • Barcode UK • Barcode Alien • Phylogeny for the UK flora • Development of bioinformatic tools • Diet analysis of endangered species • Pollinator services Thank you! • Funding from Welsh Government, National Botanic Garden of Wales, National Museum Wales, Countryside Council for Wales • Sponsorship from the people of Wales • www.gardenofwales.org.uk • Science at the Garden of Wales on Facebook