DRK12-10 Poster4_0
advertisement
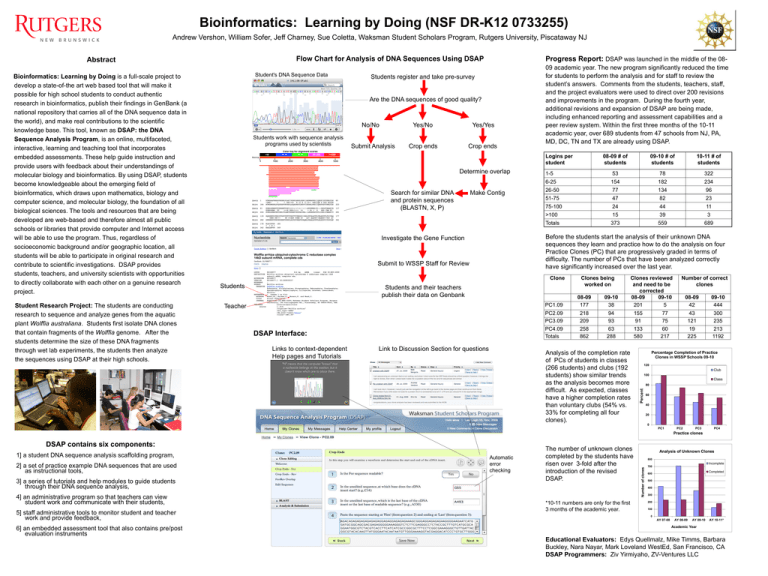
Bioinformatics: Learning by Doing (NSF DR-K12 0733255) Andrew Vershon, William Sofer, Jeff Charney, Sue Coletta, Waksman Student Scholars Program, Rutgers University, Piscataway NJ Flow Chart for Analysis of DNA Sequences Using DSAP Bioinformatics: Learning by Doing is a full-scale project to develop a state-of-the art web based tool that will make it possible for high school students to conduct authentic research in bioinformatics, publish their findings in GenBank (a national repository that carries all of the DNA sequence data in the world), and make real contributions to the scientific knowledge base. This tool, known as DSAP: the DNA Sequence Analysis Program, is an online, multifaceted, interactive, learning and teaching tool that incorporates embedded assessments. These help guide instruction and provide users with feedback about their understandings of molecular biology and bioinformatics. By using DSAP, students become knowledgeable about the emerging field of bioinformatics, which draws upon mathematics, biology and computer science, and molecular biology, the foundation of all biological sciences. The tools and resources that are being developed are web-based and therefore almost all public schools or libraries that provide computer and Internet access will be able to use the program. Thus, regardless of socioeconomic background and/or geographic location, all students will be able to participate in original research and contribute to scientific investigations. DSAP provides students, teachers, and university scientists with opportunities to directly collaborate with each other on a genuine research project. Student Research Project: The students are conducting research to sequence and analyze genes from the aquatic plant Wolffia australiana. Students first isolate DNA clones that contain fragments of the Wolffia genome. After the students determine the size of these DNA fragments through wet lab experiments, the students then analyze the sequences using DSAP at their high schools. Student's DNA Sequence Data Progress Report: DSAP was launched in the middle of the 08- Students register and take pre-survey Are the DNA sequences of good quality? No/No Students work with sequence analysis programs used by scientists Submit Analysis Yes/No Yes/Yes Crop ends Crop ends 09 academic year. The new program significantly reduced the time for students to perform the analysis and for staff to review the student’s answers. Comments from the students, teachers, staff, and the project evaluators were used to direct over 200 revisions and improvements in the program. During the fourth year, additional revisions and expansion of DSAP are being made, including enhanced reporting and assessment capabilities and a peer review system. Within the first three months of the 10-11 academic year, over 689 students from 47 schools from NJ, PA, MD, DC, TN and TX are already using DSAP. Logins per student Determine overlap Search for similar DNA and protein sequences (BLASTN, X, P) Make Contig 08-09 # of students 09-10 # of students 10-11 # of students 1-5 53 78 322 6-25 154 182 234 26-50 77 134 96 51-75 47 82 23 75-100 24 44 11 >100 15 39 3 Totals 373 559 689 Before the students start the analysis of their unknown DNA sequences they learn and practice how to do the analysis on four Practice Clones (PC) that are progressively graded in terms of difficulty. The number of PCs that have been analyzed correctly have significantly increased over the last year. Investigate the Gene Function Submit to WSSP Staff for Review Clone Students Students and their teachers publish their data on Genbank Teacher DSAP Interface: Links to context-dependent Help pages and Tutorials Link to Discussion Section for questions Clones being worked on Clones reviewed and need to be corrected 08-09 09-10 201 5 PC1.09 08-09 177 09-10 38 PC2.09 PC3.09 218 209 94 93 155 91 PC4.09 Totals 258 862 63 288 133 580 Analysis of the completion rate of PCs of students in classes (266 students) and clubs (192 students) show similar trends as the analysis becomes more difficult. As expected, classes have a higher completion rates than voluntary clubs (54% vs. 33% for completing all four clones). Number of correct clones 08-09 42 09-10 444 77 75 43 121 300 235 60 217 19 225 213 1192 Percentage Completion of Practice Clones in WSSP Schools 09-10 120 Club 100 Class 80 Percent Abstract 60 40 20 0 PC1 PC2 PC3 PC4 Practice clones 1] a student DNA sequence analysis scaffolding program, 2] a set of practice example DNA sequences that are used as instructional tools, 3] a series of tutorials and help modules to guide students through their DNA sequence analysis, 4] an administrative program so that teachers can view student work and communicate with their students, 5] staff administrative tools to monitor student and teacher work and provide feedback, 6] an embedded assessment tool that also contains pre/post evaluation instruments Automatic error checking The number of unknown clones completed by the students have risen over 3-fold after the introduction of the revised DSAP. *10-11 numbers are only for the first 3 months of the academic year. Analysis of Unknown Clones 800 Incomplete Number of clones DSAP contains six components: 700 Completed 600 500 400 300 200 100 0 AY 07-08 AY 08-09 AY 09-10 AY 10-11* Academic Year Educational Evaluators: Edys Quellmalz, Mike Timms, Barbara Buckley, Nara Nayar, Mark Loveland WestEd, San Francisco, CA DSAP Programmers: Ziv Yirmiyaho, ZV-Ventures LLC