PHYSICAL REVIEW E 111, 014414 (2025)
Editors’ Suggestion
Nonequilibrium properties of autocatalytic networks
Armand Despons
*
Gulliver Laboratory, UMR CNRS 7083, PSL Research University, ESPCI, Paris F-75231, France
(Received 7 April 2024; revised 21 October 2024; accepted 18 December 2024; published 14 January 2025)
Autocatalysis, the ability of a chemical system to make more of itself, is a crucial feature in metabolism
and is speculated to have played a decisive role in the origin of life. Nevertheless, how autocatalytic systems
behave far from equilibrium remains unexplored. In this work, we elaborate on recent advances regarding the
stoichiometric characterization of autocatalytic networks, particularly their absence of mass-like conservation
laws, to study how this topological feature influences their nonequilibrium behavior. Building upon the peculiar
topology of autocatalytic networks, we derive a decomposition of the chemical fluxes, which highlights the
existence of productive modes in their dynamics. These modes produce the autocatalysts in net excess and require
the presence of external fuel/waste species to operate. Relying solely on topology, the flux decomposition holds
under broad conditions and, in particular, does not require steady state or elementary reactions. Additionally,
we show that once externally controlled, the nonconservative forces brought by the external species do not
act on these productive modes. This must be considered when one is interested in the thermodynamics of open
autocatalytic networks. Specifically, we show that an additional term must be added to the semigrand free energy.
Finally, from the thermodynamic potential, we derive the thermodynamic cost associated with the production of
autocatalysts.
DOI: 10.1103/PhysRevE.111.014414
I. INTRODUCTION
A. Context and motivations
At the heart of cellular metabolism is its capability
to replicate essential biomolecules from preexisting ones,
employing elementary compounds as elemental building
materials. This trait emphasizes the vital role of autocatalysis, namely, systems that produce more of themselves,
in living organisms. It results in highly complex behavior [1], enabling growth [2–4] and self-reproduction [5].
These attributes are believed to have played a central role
in abiogenesis [6–13]. The growing interest in autocatalytic
networks can be traced back to the pioneering studies of
Kauffman on autocatalytic sets [14,15]. Since then, recent
studies have elucidated the topological features of autocatalytic networks, allowing classification and detection in large
networks [16,17].
Concurrently, the thermodynamics of chemical systems
has undergone intense investigations since the foundational
works of Gibbs, who introduced the concept of chemical
potential [18], and de Donder, who introduced the concept
of affinity [19], which quantifies the irreversible forces driving chemical systems. Specifically, in the 1970s, particular
attention was drawn to the stochastic dynamics of chemical
systems based on the chemical master equation [20,21]. This
has allowed for the derivation of the foundational concepts regarding nonequilibrium thermodynamics of chemical systems
[22–24]. Since then, the emergence and growing popularity
*
Contact author: armand.despons@espci.fr
2470-0045/2025/111(1)/014414(16)
of stochastic thermodynamics, combined with chemical reaction network (CRN) formalism, have led to the development
of a consistent theory assessing the nonequilibrium thermodynamics of chemical networks [25–27]. As living systems
are arguably the most important example of nonequilibrium
systems, this theory was quickly applied to study biochemical
networks [28,29].
More recently, there has been an increasing focus on establishing connections between topology and nonequilibrium
dynamics in chemical networks [30–32]. Indeed, since topology precedes dynamics, it imposes universal constraints on
the network’s behavior, which hold under a wide range of
conditions [33,34]. Yet, the connections between the specific
topology of autocatalytic networks and their nonequilibrium
behavior have not yet been explored. Therefore, this paper
aims to investigate the interplay between the topology of
autocatalytic networks and their nonequilibrium dynamics.
B. Outline of the paper
To derive our results, we build upon the stoichiometric
condition of autocatalysis given by Blokhuis et al. in Ref. [16],
recalled in Eq. (1), and we introduce the notion of autocatalytic networks. Specifically, we will consider chemical
systems where an autocatalytic network is coupled with external species acting as fuel and waste materials, serving to
produce autocatalysts in excess and allow for mass conservation in the system. These assumptions are captured by the
topology of such networks, encoded in their stoichiometric
matrices, which follow the block decomposition given in
Eq. (3).
014414-1
©2025 American Physical Society
ARMAND DESPONS
PHYSICAL REVIEW E 111, 014414 (2025)
Then, in Sec. II, by relying solely on the topology of such
networks, we derive their nonequilibrium behavior. The main
result of this section is the decomposition of the chemical
fluxes in Eq. (28) on the unique linear basis distinguishing
pathways that produce the autocatalysts in excess (productive
modes) and pathways preserving the state in the autocatalytic network (cycles). As the analysis is solely based on
stoichiometry, this decomposition is independent of the chemical fluxes. In addition, we show that the physical role of
the chemical species exchanged with the environment is fully
captured by the topology and depends on whether or not they
break a conservation law. Particularly, we demonstrate that,
even though both the productive modes and the cycles of the
autocatalytic network require influx of external species, they
are in fact two drastically different processes. We will illustrate the main ideas of this section on the network associated
with glucose metabolism and its regulation (glycolysis and
gluconeogenesis).
In Sec. III, we dwell on the thermodynamics of autocatalytic networks. Specifically, in this section we derive the
nonequilibrium thermodynamic potential of autocatalytic networks. It is obtained from the semigrand Gibbs free energy
by adding an extra term similar to the potential energy associated with conservative forces in Newtonian mechanics.
The latter represents the conservative forces that the fuel and
waste species exert on the reactions when they are converted
into an excess of autocatalysts; it is expressed using a gauge
transform of the chemical potentials. The main results of
this section are then the expression of the thermodynamic
potential in Eq. (56) and its associated entropy production rate
decomposition in Eq. (61). Finally, we focus on autonomous
networks in which the cost of sustaining the productive modes
and the cycles takes a simple form.
As a result, the stoichiometric condition prescribes that the net
change of autocatalytic species along the overall reaction, i.e.,
S · u = β − α, contains only strictly positive components:
S · u > 0,
(1)
component-wise.
For simplicity, we will assume in the main text that the
rows of S are linearly independent:
ker S = {0}.
(2)
This stronger condition is sufficient (yet not necessary) to
satisfy the stoichiometric condition in Eq. (1) and is observed
for most autocatalytic networks. We relax this assumption in
Appendices A and B to address the most general framework
of Eq. (1).
2. External species
The existence of a strictly productive reaction imposed
by the stoichiometric condition Eq. (1) violates mass conservation. Hence, the autocatalytic network cannot exist
independently and must be coupled to external species, which
also partake in the autocatalytic reactions as food/waste
species or external catalysts. In addition, the external species
might also be involved in some additional reactions. In the
following, we let E be the set of the external species. As a
result, the topology of the network formed by considering
both types of species (external and autocatalytic) is encoded
in its stoichiometric matrix, ∇, which has the following block
decomposition:
(3)
C. Setup and notations
1. Autocatalytic CRNs
We consider an autocatalytic network composed of reacting chemical species that change through chemical reactions.
We refer to the chemical species in the autocatalytic network
as the autocatalytic species, and we let Z be the set containing
all the autocatalytic species. Similarly, the reactions in the
autocatalytic network are called the autocatalytic reactions
that we gather in the set R. The net change of species z ∈ Z
along reaction ρ ∈ R is Sρz ; it is positive (resp. negative) if z
is produced in net excess (resp. consumed) by the reaction.
Subsequently, the stoichiometric matrix S = {Sρz } quantifies
the net change of autocatalytic species (the rows of S) along
all the autocatalytic reactions (the columns of S) [35]. The
autocatalytic nature of the network should be reflected in its
topology, which is encoded in the stoichiometric matrix S.
Specifically, following Blokhuis et al. [16], the stoichiometric condition for autocatalysis is the existence of a pathway
creating a net excess of all the autocatalytic species. Such a
pathway consists of a linear combination of the autocatalytic
reaction, u, which can be represented as an overall reaction,
In this decomposition, ∇ ER stands for the restriction of ∇ on
the subsets of external species (E) and autocatalytic reactions
(R), which quantifies how the external species participate in
the autocatalytic reactions. In contrast, the additional reactions (\R) involve only external species. In practice, given
a (possibly large) chemical network described by ∇, locating
an autocatalytic subnetwork amounts to finding a subset Z
of species and a subset R of reactions such that S ≡ ∇ Z
R
follows the stoichiometric condition Eq. (1) [or its stronger
counterpart Eq. (2)].
3. Dynamics
We call j (resp. v) the chemical fluxes associated with
the autocatalytic reactions R (resp. additional reactions \R).
The dynamics of the open chemical system follows the kinetic
rate equations [36] which, from the block decomposition of
the stoichiometric matrix, can be written as
u
→ β · z,
α·z−
dt [e] = ∇ ER · j + ∇ E\R · v + I E ,
(4)
where z contains the autocatalytic species, and α (resp. β)
stores the reactant (resp. product) stoichiometric coefficients.
dt [z] = S · j + I Z ,
(5)
014414-2
NONEQUILIBRIUM PROPERTIES OF AUTOCATALYTIC …
PHYSICAL REVIEW E 111, 014414 (2025)
where [s], for s ∈ S (with S = Z or E), contains the chemical
concentrations of species in the subset S, and I S contains the
external fluxes which couple the chemical species with the
environment. In what follows, we will assume that all the external species are exchanged with the environment, I e = 0 for
all e ∈ E. On the contrary, for the autocatalytic species, we let
Y ⊂ Z (resp. X ⊂ Z) be the subset of autocatalytic species
subjected to a nonvanishing (resp. vanishing) external flux,
i.e., I y = 0 for all y ∈ Y and I X = 0. As a result, the dynamics
in the autocatalytic subnetwork is
dt [y] = S Y · j + I Y ,
dt [x] = S X · j.
(6)
4. Networks of elementary reactions
In Sec. III we analyze the thermodynamics of an open autocatalytic network. In doing so, for this section, we will further
assume that the network is decomposed into elementary reactions such that, whenever a reaction exists, its backward
counterpart also exists. Hence, the stoichiometric matrix and
the elementary fluxes can be written as the result of a forward
and backward contribution:
j = j+ − j−,
∇ = ∇− − ∇+ ,
v = v+ − v−,
(7)
where ∇ − (resp. ∇ + ) describes the product (resp. reactant)
stoichiometry and j + /v + (resp. j − /v − ) are the forward (resp.
backward) unidirectional fluxes. In addition, we assume that
the stochastic trajectories in the concentration space follow
the chemical master equation. Taking the large volume limit
with fixed concentrations in the chemical master equation we
obtain the deterministic regime whose dynamics follows the
rate equations written above [20,21,37]. In this limit, the
stochastic rates peak at their most probable value, yielding
mass-action law for the unidirectional fluxes [38,39],
s
E s
j ±ρ = k ±ρ
[s](∇± )ρ , v ±r = ν ±r
[e](∇± )r , (8)
s ∈ E∪Z
e∈E
where ρ ∈ R and r ∈ \R, and with k ±ρ /ν ±r the kinetic rate
constants.
For simplicity, we will further assume that the mixture
behaves ideally such that the chemical potential of species
s ∈ Z ∪ E is
μs = μ◦s + RT ln [s].
(9)
The local detailed balance condition relates the standard
chemical potentials to the kinetic rate constants, ensuring
thermodynamic consistency:
k +ρ
ν +r
μ◦ · ∇ ρ
μ◦ · ∇ r
,
ln
,
(10)
=
−
=
−
k −ρ
RT
ν −r
RT
where ∇ ρ (resp. ∇ r ) corresponds to the column associated
with the autocatalytic reaction ρ ∈ R (resp. additional reaction r ∈ \R) in the full stoichiometric matrix.
ln
II. NONEQUILIBRIUM AUTOCATALYTIC NETWORKS
A. Conservation laws
1. Definition
Conservation laws of a CRN are the row vectors that
belong to the left null space of the full stoichiometric matrix
[25,27,40],
· ∇ = 0.
(11)
Indeed, when the CRN is closed, i.e., when I = 0, L = · [s]
is a conserved quantity: dt L = 0. Among the conservation
laws, those that have only positive entries, s ⩾ 0 for all
species s, are called mass-like conservation laws. Lavoisier’s
law of mass conservation implies that the subset of mass-like
conservation laws is never empty. In addition, there always
exists a conservation law such that s > 0 for all species
s, expressing total mass conservation in the closed system.
While conservation laws are generally difficult to interpret,
moieties form a subset of mass-like conservation laws that
can be understood as the molecular fragments exchanged between species during chemical reactions [41]. For instance,
in biochemical networks, common moieties include carbon
conservation and phosphate group conservation [see also
Fig. 1(a)].
2. Moiety matrix
As a result, our stricter condition for autocatalysis Eq. (2)
amounts to the absence of conservation laws in the subnetwork. The absence of conservation laws in S implies that they
must be brought by the external species E. We can consider
a linear basis of the conservation laws that we index by i,
{i }. We then define the matrix L = {i }, where the ith row
represents the conservation law i . As a result, L is a full-rank
matrix satisfying L · ∇ = 0. Consequently, all the mass-like
conservation laws of ∇ are either a row of L or can be
obtained as a linear combination of the rows of L.
From the block decomposition Eq. (3), we have
LE · ∇ ER + LZ · S = 0,
LE · ∇ E\R = 0.
(12)
Now, as the rows of L are linearly independent, there exists a
subset of external species E p ⊂ E such that the restriction of L
to this subset, LE p , is (square) nonsingular. Additionally, we
denote E f ⊂ E as the remaining external species: E = E p ∪
E f . Splitting Eq. (12) on these new subsets yields
E
E
LE p · ∇ Rp + LE f · ∇ Rf + LZ · S = 0,
(13)
and similarly for the additional reactions \R. Then, from the
nonsingularity of LE p , we have
E
E
∇ Rp = −ME f · ∇ Rf − MZ · S,
(14)
where we introduce the moiety matrix of species E p [27],
−1
M = LE p
· L = 1|E p | , ME f , MZ ,
(15)
in which 1|E p | stands for the |E p | × |E p | identity matrix. The
moiety matrix is the row-reduced form of L, which defines
another linear basis for the conservation laws: M · ∇ = 0.
Thus, with Eq. (15), it becomes explicit that each conservation
law (i.e., each row of L) is paired with a single species in
E p such that setting a nonvanishing external flux on species
e p ∈ E p (I e p = 0) breaks its associated conservation law in
L. When the ith row of L, i , corresponds to a moiety, its
associated species in E p represents the exchanged fragment,
and the nonvanishing entries in the ith row of M indicate
where this fragment appears in the other species. However, for
014414-3
ARMAND DESPONS
(a)
PHYSICAL REVIEW E 111, 014414 (2025)
(b)
(c)
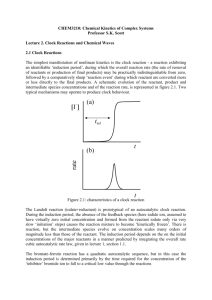
FIG. 1. Stoichiometric autocatalysis in glucose metabolism. (a) Simplified representation of glucose degradation into pyruvate (glycolysis)
and its regulatory pathway (gluconeogenesis) adapted from Ref. [43] (Chap. 14, Fig. 16). In this network, the autocatalytic species Z are
represented in the right angle boxes while the external species E are in the broken angle boxes. For simplicity, we did not balance the reactions
in H+ and H2 O and we omitted the enzymes catalyzing the reactions. (b) There exists a linear combination of the reactions, u, that produces
an excess of all the autocatalytic species Z at the expense of the external species, ensuring the existence of an autocatalytic subnetwork in
the glucose metabolism. (c) The external potential species E p (black broken angle boxes) can be paired with the conservation law that they
break once externally controlled. In particular, carbon and phosphate group exchange among the species are two important moieties in the
metabolism and are here represented using the color level and the purple octagons, respectively. The remaining external species (ATP and
GTP) are the external force species E f that bring nonconservative forces that break detailed balance.
conservation laws that cannot be interpreted as an exchanged
fragment, the nonvanishing entries in M generalize the notion
of a moiety.
3. Broken conservation laws
Once all external species are coupled to the environment,
their physical roles depend on whether they belong to E p or
to E f . Each species in E p breaks its associated conservation
law, while species in E f induce flows through the system,
effectively connecting external species. As a result, species
E p remove constraints from the system, while species E f enforce nonvanishing fluxes through the reactions. From this,
we can anticipate that if only potential species are present,
the system cannot sustain nonvanishing fluxes, which prevents
the network from settling into a nonequilibrium steady state
(NESS). On the other hand, species E p are referred to as the
(external) potential species and imposing a nonvanishing flux
on all the species in E p breaks all the conservation laws of the
network. Conversely, the species in E f are referred to as the
external force species: once coupled to the environment, they
are capable of breaking detailed balance, thereby maintaining
nonvanishing fluxes through the open system [27,42].
From the stronger stoichiometric condition Eq. (2), the
autocatalytic subnetwork lacks conservation law. Hence, once
the external species are exchanged with the environment by
the means of external fluxes, the rows of L provide the broken
conservation laws of the open system, which play a pivotal
role in its thermodynamics.
Remark. When the autocatalytic subnetwork follows only
the most general criterion Eq. (1) instead of Eq. (2), it is
instead deprived of mass-like conservation laws. In that case,
all the conservation laws of the subnetwork have entries of
opposite sign [see also Eq. (A1) in Appendix A]. In that
case, there might also be broken conservation laws in the
subnetwork originating from the autocatalytic species that are
externally controlled, i.e., the Y species. When this arises,
one should further split the subset of externally controlled
autocatalytic species Y into potential species, Y p , breaking the
conservation laws of the subnetwork, and force species Y f .
014414-4
NONEQUILIBRIUM PROPERTIES OF AUTOCATALYTIC …
PHYSICAL REVIEW E 111, 014414 (2025)
4. Elementary modes of production
From the stricter condition for autocatalysis in Eq. (2),
the rows of S are linearly independent; thus S admits a
(nonunique) right inverse G:
S · G = 1|Z| .
(16)
The column associated with species z ∈ Z in G is denoted
gz . It defines a pathway that produces one unit of species z
in excess, while leaving the other autocatalytic species unaffected. Consequently, we label gz as the elementary mode
of production of species z, and, subsequently, the columns
of G are the productive modes of the autocatalytic subnetwork. Considering the sum of all the elementary modes, u =
z∈Z gz , defines a pathway that produces one unit of all the
autocatalytic species in excess:
S · u = 1 > 0.
(17)
Hence, as claimed in the introduction, the stricter condition
for autocatalysis is sufficient to ensure that the stoichiometric
criterion in Eq. (1) holds. Finally, as S is integer-valued, the
productive modes can always be rescaled, G → nG for n ∈
N ∗ , so that all the elementary modes are also integer-valued.
Doing so, the (rescaled) elementary modes produce an excess
of n units of each autocatalytic species.
Remark. When the autocatalytic subnetwork follows the
most general criterion of Eq. (1), the presence of (nonmass-like) conservation laws prevents the definition of an
elementary mode for each of the autocatalytic species. Instead, only a subset Z f of autocatalytic species will be
associated with the elementary modes, and the production of
the other autocatalytic species (Z p = Z − Z f ) will be subjected to the production of species in Z f .
species P is controlled, P is broken. Another conservation
law, Glu , carbon conservation, is also illustrated by the color
background in the boxes of the species in Fig. 1(a). It is broken
once glucose is controlled. Additionally, the total conservation
of ATD/ADP, GTP/GDP, and NADH/NAD is represented by
ADP , GDP , and NAD . These are broken once APD, GDP,
and NAD are externally controlled. The last conservation law
of the network is
(18)
NADH = 1, 2, 2, 2, 2, 1, 1
NADH Glu G6P F6P F1,6P G3P DHAP
(the entries associated with the missing species are zeros) and
has no simple interpretation in terms of moiety, even though
it is mass-like. It is broken once NADH is exchanged with
the environment. The conservation laws define the rows of
the matrix L as illustrated in Fig. 1(c) [see also Eq. (D2)
in Appendix D], and the potential species, which break the
conservation laws, are
E p = {Glu, P, ADP, GDP, NAD, NADH}.
The remaining external species define the external force
species, E f = {ATP, GTP}, and are represented with blue
boxes in Fig. 1(a).
C. Cycles
1. Cycles of the network and the subnetwork
Cycles of the full CRN are the column vectors belonging to the right null space of the full stoichiometric matrix:
∇ · γ = 0. According to the block decomposition in Eq. (3)
we have
B. Application to glucose metabolism
∇ ER · γ R + ∇ E\R · γ \R = 0,
(19)
S · γ R = 0,
(20)
E = {ATP, ADP, GTP, GDP, NADH, NAD, P, Glu}.
where γ R stands for the restriction of the cycle on the subset
of reactions R = R or \R. Further, from Eq. (20), we see
that if γ is a cycle, then either γ R = 0, or γ R is itself a
cycle of the autocatalytic subnetwork and it belongs to the
right null space of S. From the rank nullity theorem, there are
| ker S | = |R| − |Z| independent cycles in the subnetwork.
In what follows, we let {cε }1⩽ε⩽|R|−|Z| be a linear basis of
these cycles. As we did for the conservation laws, we define
the matrix
The external species are represented with broken angle boxes
in Fig. 1(a), while the autocatalytic species are
C = {cε }1⩽ε⩽|R|−|Z| ,
As an example, we consider in Fig. 1(a) the network
associated with glucose metabolism [see also Eq. (D1) in
Appendix D for the stoichiometric matrix]. In the latter, glycolysis converts glucose into pyruvate with reactions 1 to 7.
On the other hand, when pyruvate accumulates, gluconeogenesis allows one to reform glucose from pyruvate with reactions
8 to 11. In this network, the external species are
Z = {G6P, F6P, F1,6P, DHAP, G3P, PEP, Pyr, Oxa},
and are represented with right angle boxes in Fig. 1(a). All
the reactions in Fig. 1(a) belong to the set R. As a result,
there exists a linear combination of autocatalytic reactions u
that produces all the autocatalytic species from the external
species, as represented in Fig. 1(b).
In this network, there are 6 independent conservation laws
which are all mass-like, while on the other hand, the autocatalytic subnetwork follows our stricter condition and is
deprived of conservation laws. A linear basis of the conservation laws can be constructed such that one, P , represents
the phosphate group exchanged between the species [represented with purple octagons in Fig. 1(a)]. Once the external
whose εth column is cε . Subsequently, the matrix C is full
rank and verifies S · C = 0.
There exists a subset R1 ⊂ R of autocatalytic reactions such that the restriction SR1 is (square) nonsingular, while R2 = R − R1 stores the remaining autocatalytic
reactions. By construction, |R1 | = |Z| while |R2 | = |R| −
|Z| = | ker S |; hence, each reaction in the subset R2 can be
paired with one cycle of the subnetwork such that removing
one reaction in R2 from the network causes precisely one
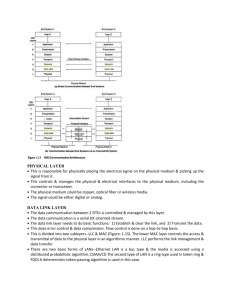
cycle of S to disappear. Physically, removing one reaction that
belongs to R2 removes exactly one cycle from the subnetwork
[see also Fig. 2(a)]. Subsequently, upon removing the reactions in R2 from the hypergraph of the network, one ends up
with the network described by SR1 , which is then deprived of
014414-5
ARMAND DESPONS
PHYSICAL REVIEW E 111, 014414 (2025)
(a)
R2
c1
R1
D. Dynamics
(b)
c2
1. Linear basis of the elementary fluxes
R1
FIG. 2. Graph-theoretic interpretation of the subsets R1 and R2 .
(a) Each cycle c of a graph is associated with one reaction in the subset R2 (represented in orange) such that, by removing this reaction,
the cycle c is removed as well. (b) Removing all the reactions R2
results in a graph deprived of cycles where the number of vertices
corresponds to the number of edges.
The right inverse G and the cycles C describe complementary effects occurring in the autocatalytic subnetwork.
Indeed, the columns of G define the productive modes of the
subnetwork that change the concentrations of the autocatalytic
species. In contrast, the columns of C are the pathways preserving the state of the subnetwork. To reflect this, we seek
for productive modes that never proceed along the reactions
in R2 by imposing G R2 = 0. This constraint removes the
nonuniqueness of G as the only right inverse fulfilling this
property is
any cycle [see also Fig. 2(b)]. Finally, the submatrix C R1 is
directly related to C R2 :
−1
C R1 = − SR1
· SR2 · C R2 .
(21)
As the columns of C are linearly independent, this relation
implies that C R2 is a (square) nonsingular matrix.
(26)
On top of that, we notice that the matrix (G, C) has a nonvanishing determinant:
2. Emergent cycles
We now examine the condition upon which a cycle of S
is also a cycle when one considers the external species. Indeed, a cycle of the subnetwork will consume and/or produce
external species. However, when a cycle of the subnetwork
preserves also the external species one has
∇ ER · c = ∇ ER1 · cR1 + ∇ ER2 · cR2 = 0.
(22)
Furthermore, we can use Eq. (21) to replace cR2 such that
Eq. (22) becomes
−1
∇ ER2 − ∇ ER1 · SR1
· SR2 · cR2 = 0.
(23)
(27)
Hence, the productive modes drawn from G in Eq. (26) and
the cyclic pathways in C define a linear basis of the flux space.
2. Decomposition of the fluxes
The chemical fluxes of the autocatalytic reactions, j, can
be decomposed on the linear basis defined by the columns of
(G, C):
In the left-hand side we recognize the Schur complement that
we will denote
ER ≡ ∇ ER − ∇ ER · SR1 −1 · SR2 .
∇
(24)
2
2
1
As a result, the right null space of the Schur complement,
E , contains all of the cycles of the subnetwork that
ker ∇
R2
E |
also preserve the external species, and there are |ker ∇
R2
independent such cycles. When all the cycles of S preserve
also the external species,
ker S = ker ∇ R ,
j = C·J+G·J
J ε (t ) cε +
=
ε
J z (t ) gz ,
(29)
z∈Z
where J ε (t ) stands for the macroscopic flux along the cycle cε
and J z (t ) is the macroscopic flux along the elementary mode
gz (see Fig. 3). Furthermore, the constraint on the right inverse
implies that the restriction of the fluxes on the subset of reactions R2 stems uniquely from the cycles of the subnetwork:
j R2 = C R2 · J =
(25)
E · C R2 = 0. From the nonsingularity of C R2 ,
one has ∇
R2
this occurs if, and only if, the Schur complement vanishes,
E = 0.
∇
R2
In contrast, the cycles of the subnetwork that are not also
cycles of the full network consume external species without
production of autocatalytic species. In the CRN literature,
they are labeled emergent cycles as they exist only once all
the external species are externally controlled. In metabolic
networks, emergent cycles are also referred to as futile cycles
because they are useless for producing metabolites and are
energy consuming (see next section).
(28)
ε
2
J ε (t ) cR
ε .
(30)
Hence, the macroscopic fluxes along the cycles of S are
completely determined by the elementary fluxes of reactions
in R2 :
−1 R2
J = C R2
·j .
(31)
In contrast, the macroscopic fluxes in J represent the rate at
which the autocatalytic species are produced or consumed in
the subnetwork. Precisely, if the macroscopic flux associated
with z ∈ Z is positive (resp. negative), J z > 0 (resp. J z < 0),
the concentration of species z increases (resp. decreases) in the
014414-6
NONEQUILIBRIUM PROPERTIES OF AUTOCATALYTIC …
PHYSICAL REVIEW E 111, 014414 (2025)
Hence the uptake of external species needed for producing an
excess z = [z](t ) − [z](0) of autocatalytic species is
e(z) = ∇ ER · G · z,
(37)
which depends only on the excess produced. In contrast, the
second term on the right-hand side of Eq. (35) quantifies the
uptake of external species along the cycles of the subnetwork.
Using Eq. (21), it can be written with the Schur complement:
ER · C R2 · J.
∇ ER · C · J = ∇
2
FIG. 3. Schematic representation of an open autocatalytic network described by Eq. (3). To fulfill mass conservation, the
autocatalytic subnetwork, represented inside the right angle box,
must be coupled with external species. The autocatalytic reactions,
R, split into two disjoint subsets. The reactions in R1 convert e(z)
external species E into an excess of autocatalytic species Z, while
the reactions in R2 are associated with the cycles of S. These are
sustained thanks to a nonconservative influx of external species. The
external species and the Y species are exchanged with the environment (delimited by the gray box) via external fluxes. Exchanging
the Y species with the environment drives nonzero fluxes along
their respective elementary modes, promoting their production (or
consumption) in the system.
subnetwork as its elementary mode, gz , is performed forward
(resp. backward).
Unlike the macroscopic fluxes along the productive modes,
J cannot be solely expressed as a function of j R1 . Indeed,
even though the fluxes along the cycles of S can be expressed
solely with j R2 via Eq. (30), part of j R1 is also due to the
cycles. Specifically, from Eq. (21),
−1
−1
− SR1
· SR2 · C R2 · J = − SR1
· SR2 · j R2 (32)
is the contribution of the cycles in j R1 . As a consequence, J
is obtained by removing this contribution:
J = SR1 · j R1 + SR2 · C R2 · J = S · j.
(33)
3. Influx of external species
Injecting the decomposition of the fluxes into the kinetic
rate equations of the autocatalytic species provides
dt [z] = J + I Z .
(34)
Similarly, by injecting the decomposition of the fluxes into the
rate equations of the external species we obtain
dt [e] = ∇ ER · G · J + ∇ ER · C · J + ∇ E\R · v + I E . (35)
The first term on the right-hand side corresponds to the influx
of external species needed to produce (or consume) autocatalytic species using the productive modes; using Eq. (34) it
can be expressed as
∇ ER · G · J = dt e(z) − ∇ ER · G · I Z .
(36)
(38)
Because the cycles of the subnetwork that are also cycles
of the full network belong to the right null space of the
Schur complement, these cycles do not contribute in Eq. (38)
where only the emergent/futile cycles remain. In particular,
if ker S = ker ∇ R , all the cycles of the subnetwork are also
preserving the external species and, in that case, the Schur
complement vanishes, and so it is for Eq. (38). Finally, the
last two terms in Eq. (35) are, respectively, the change of
external species along the additional reactions (\R) and the
fluxes coupling the external species with the environment.
From Eq. (36) we see that the rate at which the external
species are injected/released along the productive modes of
the autocatalytic subnetwork can be written as an exact time
derivative. By integrating this term we have expressed the uptake of external species to produce the autocatalytic species in
Eq. (35). The latter is independent of the pathway taken in the
subnetwork and depends only on the excess of autocatalytic
species produced (or consumed). Reminiscent of Newtonian
mechanics, we now refer to the influx of external species
along the productive modes as the conservative influx. As we
shall see in the next section, this analogy can be pushed further
as the chemical forces driving this process can be derived from
a potential energy.
On the contrary, the influx of external species along the
cycles of the subnetwork cannot be expressed as an exact
time derivative, and, as a result, they must be sustained by
the means of nonconservative forces.
4. Steady fluxes
From Eq. (6), steady state in the open subnetwork is
achieved when the concentrations of the autocatalytic species
that are not externally controlled, i.e., the X species, become
constant in time. This situation occurs when the fluxes verify
j ∈ ker S X [29,44]. Clearly, all the cycles of S remain cycles
for S X and, then, the columns of C define a family of linearly
independent vectors generating a subset of ker SX . Additionally, by definition the elementary modes of the species Y leave
each of the X species unaffected. As a result, the columns of
GY also generate a subset of ker SX and are linearly independent from the columns of C. Finally, the rank-nullity theorem
implies that the columns of (C, GY ) form a linear basis of
ker S X . Then, at steady state, the decomposition of the fluxes
becomes
j = C · J + GY · J Y .
(39)
Removing or adding intermediate species involved in unimolecular reactions (fueled with an arbitrary number of
external species as reactants and products) is an important
class of coarse graining which preserves the decomposition
014414-7
ARMAND DESPONS
PHYSICAL REVIEW E 111, 014414 (2025)
of the steady fluxes. This procedure is, for example, used
to derive Michaelis-Menten kinetics or Hill kinetics from
elementary reactions [45]. Aside from linear pathways, more
advanced structural reductions which do not affect steady state
have been recently studied in Ref. [46]. Finally, the decomposition of the steady fluxes serves as the starting point to
construct a circuit theory of open chemical networks, drawing
an analogy between CRNs and electronic circuits [47,48].
5. Physical role of the species
As a direct consequence of Eq. (39), when only the external species are externally controlled (Y = ∅), the steady
fluxes align with the cycles of the subnetwork. Among these
cycles, only the emergent cycles of the subnetwork consume
external species and, then, the nonvanishing contributions in
Eq. (38) stem only from the emergent cycles of the autocatalytic subnetwork. Hence, maintaining nonzero fluxes along
the emergent cycles requires one to exert nonconservative
forces on the system. Additionally, when only the external
species are controlled, only the external force species E f bring
such forces in the network. Hence, we can relate the number
of external force species, |E f |, to the number of emergent
cycles in the autocatalytic subnetwork (see Appendix C for
the derivation):
ER .
|E f | = |\R| + |R2 | − ker ∇
2
(40)
From that the number of conservation laws brought by the
external species follows:
|E p | = |E| − |E f |.
(41)
In other words, Eq. (40) states that the physical role of the
external force species in the open system is (i) to maintain all
the additional reactions away from equilibrium and (ii) to sus E | emergent
tain nonvanishing fluxes along the |R2 | − |ker ∇
R2
cycles of the subnetwork.
Exerting nonzero external fluxes on the Y species causes
|Y| new cycles to exist for the remaining X species. These
are all emergent as they do not exist prior to controlling the
Y species. On the other hand, as they do not break any conservation law, the Y species are “force species” that maintain
the fluxes along their respective elementary modes of production. Indeed, to sustain steady production (or consumption) of
species y, the latter must be coupled with the environment by
the means of a nonvanishing external flux, I y = 0, such that
J y (t ) −→ J y = −I y (see also Fig. 3). Finally, the macroscopic fluxes along the X species tend to vanish as NESS
settles in the subnetwork, J X (t ) −→ 0. Nevertheless, during
the transient dynamics, J X are nonvanishing and serve as the
relaxation modes of the subnetwork to reach steady state [30].
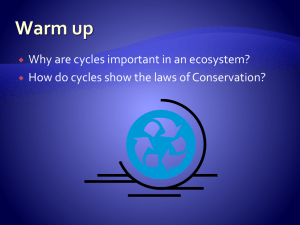
where the entries in the remaining reactions vanish. These cycles are caused by the inhibitory reactions of gluconeogenesis,
R2 = {9, 10, 11}, which are represented with orange arrows
in Fig. 4(a). Removing these reactions results in a network
deprived of cycles, where only the reactions in R1 remain [red
arrows in Fig. 4(a)]. These three cycles define the columns of
the matrix C. Taking the external species into account, the
full network has one cycle γ [gray dashed arrows in Fig. 4(a)]
that is formed by a linear combination of two cycles of the
subnetwork:
γ = c1 − c2 .
(43)
Hence, the kernel of the Schur complement is of dimension
1 [see also Fig. 4(b)]. Thus, when glycolysis and gluconeogenesis are considered simultaneously, two independent
emergent/futile cycles exist. These two degrade ATP/GTP
into ADP/GDP without any useful production of metabolite:
c1 &c2
ATP −−−→ ADP + P,
c3
GTP −
→ GDP + P.
On the other hand, the reactions in R1 are sufficient to define
the elementary modes of the autocatalytic species that are
linearly independent of the three cycles of the subnetwork.
These elementary modes are the columns of the matrix G:
each column of G defines an overall reaction that produces an
excess of its associated autocatalytic species; see Fig. 4(c).
Consequently, the fluxes of the autocatalytic reactions, j,
can be written on the basis defined by (C, G) according to
Eq. (28). When pyruvate is externally controlled, the dynamics tends to align the fluxes along its mode, gPyr , such that, at
steady state,
3
J ε cε + J Pyr gPyr .
j =
(44)
ε=1
This results in the well-known conversion of one glucose unit
into two units of pyruvate that replenishes ATP from ADP, as
represented in Fig. 4(d).
Here, one cycle is shared between the autocatalytic subnetwork and the full network (γ = c1 − c2 ); hence Eq. (40)
imposes that there should be two external force species in
order to maintain the fluxes along the emergent cycles of the
subnetwork. Finally, we note that the mode of production of
DHAP requires performing reaction 5 backward. In this case,
it is admissible as this reaction is indeed reversible. If this
were not the case, the macroscopic flux along this mode would
always vanish.
III. THERMODYNAMICS OF AUTOCATALYTIC
NETWORKS
E. Application to glucose metabolism II
A. Fundamental nonconservative forces
In glucose metabolism, there are three more reactions than
the number of autocatalytic species (|R| = 11 and |Z| = 8);
as a result, the rank-nullity theorem imposes that there are
three independent cycles in the autocatalytic subnetwork:
The total entropy production rate (EPR) quantifies the
amount of energy dissipated by the reactions in the system.
It can be expressed as the contribution of the reactions of the
subnetwork (R) and the additional reactions (\R):
c1 = (1,
1
1) ,
11
c2 = (1,
3
1) ,
10
˙ =
˙R +
˙ \R ⩾ 0.
c3 = (1,
1,
1) ,
7
8
9
(42)
Starting from the chemical master equation that governs the
stochastic trajectories in the concentration space, and taking
014414-8
NONEQUILIBRIUM PROPERTIES OF AUTOCATALYTIC …
(a)
PHYSICAL REVIEW E 111, 014414 (2025)
(b)
(c)
(d)
FIG. 4. Nonequilibrium dynamics of glucose metabolism. (a) The reactions associated with glycolysis and the conversion of pyruvate into
oxaloacetate of gluconeogenesis (reaction 8) define the subset R1 , represented with solid red arrows. The other reactions of gluconeogenesis
bring cycles in the subnetwork and define the subset R2 , represented with solid orange arrows. (b) A linear basis of the cycles of the subnetwork
is formed by these three cycles. Conversely, the full network has only one cycle [represented with the gray dotted lines in (a)], formed by a
linear combination of two cycles of the subnetwork. As a consequence, the cycles shared by the full network and the subnetwork are a linear
space of dimension 1. (c) The subset R1 allows one to define independent pathways producing each of the autocatalytic species that are
linearly independent from the cycles. (d) Setting a nonvanishing external flux on the pyruvate one selects its elementary mode at steady state,
recovering the conversion of one unit of glucose into two units of pyruvate that regenerates ATP from ADP [49].
the large volume limit, ends up in the following form for
the dissipation of the dissipation along the reactions of the
subnetwork in the deterministic regime [38],
˙R =
T
(j
+ρ
−j
−ρ
) ln
ρ∈R
j +ρ
,
j −ρ
(45)
expressed in units where RT = 1. In the dissipation, the log
ratio of the unidirectional fluxes defines the chemical forces
acting on the reactions, i.e., the affinities
Aρ ≡ ln
j +ρ
j −ρ
= −(μ · ∇)ρ = −Gρ ,
(46)
such that the affinities of the reactions can be written as
Aρ = −(F · ∇)ρ .
(48)
As a result, when F = 0, all the affinities vanish implying that
the subnetwork has reached detailed balance. Hence, the entries in F are the fundamental nonconservative forces [27,40]
associated with each of the chemical species. As anticipated,
by definition, these forces necessarily vanish for the potential
species: F E p = 0.
Finally, the dissipation along the autocatalytic reactions
can be written as a function of the nonconservative forces of
the species:
˙ R = −F · ∇ R · j.
T
(49)
which correspond to the opposite of the Gibbs free-energy differences of the reactions. Using Eq. (14) allows us to replace
the stoichiometric matrix of the external potential species,
E
∇ Rp , in Eq. (46). By doing so, we introduce the following
gauge transform of the chemical potentials:
Similarly, the dissipation along the additional reactions reads
F = μ − μE p · M,
From the decomposition of steady fluxes Eq. (39), when
autocatalytic species are exchanged with the environment
(47)
˙ \R = −F · ∇ \R · v = −F E · ∇ E\R · v.
T
(50)
B. Unconditionally detailed balanced subnetworks
014414-9
ARMAND DESPONS
PHYSICAL REVIEW E 111, 014414 (2025)
(Y = ∅), macroscopic currents flow along the productive
modes of the Y species at steady state, such that reaching
detailed balance (equilibrium) in the subnetwork is impossible. In contrast, when Y = ∅, the subnetwork might still be
able to reach equilibrium for arbitrary values of the chemical
potentials of external species μE . When this occurs, the subnetwork is said to be unconditionally detailed balanced [27]
and the chemical potentials of the free autocatalytic species,
μX = μZ , are able to balance arbitrary μE :
way as potential energies in Newtonian mechanics, namely
“the force times the displacement.”
E
μeq
Z · S = −μE · ∇ R .
Gathering the terms on the right-hand side of Eq. (57) as
follows,
As SR1 is nonsingular, the unique candidate for μeq
Z is
−1
E
μeq
.
Z = −μE · ∇ R1 · SR1
(51)
D. Decomposition of the EPR
The time evolution of G can be written as
˙ − dt μE · m + dt F E · e
dt G = − T p
+ F E · (I E + dt e) + F Z · I Z .
(52)
Plugging this candidate in the equations associated with the
reactions in R2 provides the following feasibility condition:
ER = 0.
μE · ∇
2
(53)
In light of the previous chapter, unconditional detailed balance
is achieved if, and only if, the Schur complement vanishes,
i.e., when
ker S = ker ∇ R .
Importantly, this condition is independent of the presence of
nonconservative forces brought by the external force species,
F E f , and relies solely on topology. This is expected from
Eq. (40) because, when the Schur complement vanishes, the
external force species serve only to maintain nonzero fluxes
along the additional reactions \R.
e∈E
[z](μz − 1),
where the “−1” is the contribution of the solvent. By setting
off external fluxes, the system starts exchanging moieties with
its environment:
(55)
In the open system,
G(z, e) = G(z, e) − μ p · m(z, e) + F E · e(z)
Ẇnc,E = F E · (I E + dt e),
(59)
Ẇnc,Z = F Z · I Z ,
(60)
(56)
corresponds to the thermodynamic potential of the open subnetwork which captures its thermodynamics behavior. The
first two terms in the definition of G correspond to the
semigrand free energy of an open CRN which was derived
previously for stochastic [40] or deterministic open chemical networks [25,27,42]. Yet, the semigrand free energy fails
to predict relaxation toward equilibrium for unconditionally
detailed balanced subnetworks in the presence of additional
reactions. To fix this, we should to take into account that the
nonconservative forces of the external species are not exerted
along the productive modes of the subnetwork because the
influx of external species along the productive modes of the
subnetwork is conservative. This is implemented by the last
term in Eq. (56), F E · e, which is expressed in the same
(61)
In this decomposition, Ẇdriv represents the driving work rate
required to change the equilibrium state in the autocatalytic
subnetwork by varying the chemical potentials of the potential
species. Note that, because the equilibrium state depends on
all the external species, the external force species also contribute to the driving work. The two nonconservative work
rates, Ẇnc, E and Ẇnc, Z , describe the power performed by
species E and Z to maintain nonvanishing fluxes. Hopefully,
the potential species does not contribute to Ẇnc, E .
E. Autonomous autocatalytic network
When the driving rate vanishes,
Ẇdriv = 0,
(54)
z∈Z
m(z, e) = ME · [e] + MZ · [z].
(58)
˙ + Ẇdriv + Ẇnc, E + Ẇnc, Z .
dt G = −T The Gibbs free energy of the closed system (I = 0) is well
known:
[e](μe − 1) +
Ẇdriv = −dt μE p · m + dt F E · e,
allows us to write the change in free energy in a thermodynamically appealing way,
C. Thermodynamic potential
G(z, e) =
(57)
(62)
the subnetwork is said to be autonomous. This is implemented
by fixing the chemical potentials of the external species to a
constant value in the system through external particle reservoirs (chemostats). Autonomy can also be achieved if there
exists a timescale separation between the dynamics of the
external species and the one of the autocatalytic subnetwork
such that, in the timescale of the subnetwork, the chemical
potentials of the external species can be regarded as constant.
In that case, the external fluxes on the external species I E
balance the consumption/production of external species by
the chemical reactions in Eq. (35).
1. Thermodynamic cost of production
For autonomous subnetworks, the decomposition of the
EPR can be written as
˙ R + Ẇcyc + Ẇprod .
dt G = −T (63)
In this decomposition,
ER · C R2 · J
Ẇcyc = −F E · ∇
2
(64)
accounts for the power needed to sustain the emergent/futile
cycles of S. Furthermore, the total work rate sustaining the
cycles can be further decomposed on each cycle cε of the
014414-10
NONEQUILIBRIUM PROPERTIES OF AUTOCATALYTIC …
PHYSICAL REVIEW E 111, 014414 (2025)
subnetwork. Indeed, as from Eq. (48) −F · ∇ = A, the εth
entry in
potential,
G = G(z, e) − G(zeq , e)
ER · C R2 = −F · ∇ R · C = A · C
−F E · ∇
2
=
is A[cε ] = −G[cε ], the overall affinity associated with cycle
cε which is the opposite of its free-energy difference. As a
result, the power maintaining the cycles can be written as
A[cε ] J ε = −
Ẇcyc =
G[cε ] J ε ,
ε
(65)
ε
such that the εth summand corresponds to the specific power
allocated to cycle cε , which vanishes if cε is also a cycle of
the full network because its affinity vanishes in that case.
Consequently, only the emergent/futile cycles will require
energy to be maintained while producing no net excess of
autocatalytic species.
In addition,
−1
Ẇprod = F E · ∇ ER1 · SR1
+ F Z · IZ
(66)
represents the power needed to sustain the production of the
autocatalytic species Z along their elementary modes. Its first
term,
−1 Z
·I ,
F E · ∇ ER1 · SR1
corresponds to the uptake of external species fueling the elementary modes of production. As the flux decomposition
suggested, the production of the autocatalytic species is driven
by the external fluxes on the Y species:
Ẇprod = −
A[gy ] I y =
G[gy ] I y ,
(67)
A[gy ] = −F · ∇ R · gy = −F E · ∇ ER · gy − Fy
(68)
y∈Y
y∈Y
where
is the overall affinity along the elementary mode of species y,
gy , which is the opposite of its Gibbs free-energy difference
G[gy ]. Similarly to the cycles, the yth summand in Eq. (67)
is the specific work rate dedicated to the production of
species y.
z∈Z
E
R
=0
∇
2
and
Y = ∅.
When these two conditions are fulfilled, the subnetwork is
unconditionally detailed balanced and the optimum of G with
respect to autocatalytic species that are not subjected to an
external flux, i.e., species in X = Z, corresponds to
−1
E
,
(69)
μeq
Z = −μE · ∇ R1 · SR1
which is in accordance with what we found by balancing
the chemical potentials in Eq. (52). Furthermore, the state
in Eq. (69) is the global minimum of the thermodynamic
[z]
[z]eq
− ([z] − [z]eq )
= L(z zeq ) ⩾ 0,
(70)
in which we expressed the relative entropy
between two nonnormalized distributions as L(a b) = i ai ln(ai /bi ) − (ai −
bi ). The latter is positive and vanishes if, and only if, the
two distributions are identical. Finally, the convexity of the
thermodynamic potential derives from the convexity of the
relative entropy [50].
3. Hill-Schnakenberg decomposition
By injecting the decomposition of the elementary fluxes
˙ R = −F · ∇ R · j, we recover the Hillin the EPR, T Schnakenberg decomposition of the EPR [22,24]:
˙ R = −F E · ∇ · C · J − F · ∇ R · G · J
T
A[cε ] J ε +
=
ε
A[gz ] J z
z∈Z
˙ cyc + T ˙ prod ,
= T
(71)
where, as before, A[gz ] denotes the overall affinity along the
elementary mode gz , and A[cε ] the overall affinity along the
cycle cε (which vanishes if cε is also the cycle for the external
species). Importantly, the dissipation along the cycles of the
˙ cyc , corresponds to Ẇcyc in Eq. (65). Subsesubnetwork, T quently, the power maintaining the emergent/futile cycles of
the subnetwork ends up dissipated. On the other hand, the
˙ prod , differs from
dissipation along the productive modes, T Ẇprod , allowing for work extraction and free-energy change in
the subnetwork. Finally, at steady state,
A[cε ] J ε +
˙R =
T
ε
A[gy ] J y .
(72)
y∈Y
Because at steady state, the production of the autocatalytic
species must be balanced by the external fluxes, J y = −I y ,
one recovers that once NESS is settled, all the power ends up
dissipated,
2. Unconditionally detailed balanced subnetworks
From the EPR decomposition in Eq. (63), it is clear that
whenever Ẇcyc = 0 and Ẇprod = 0, the thermodynamic po˙ R ⩽ 0. These
tential G tends to its minimum: dt G = −T conditions are equivalent to
[z] ln
˙ R = Ẇ cyc + Ẇ prod ,
T
(73)
and the free energy is constant in time dt G = 0.
IV. CONCLUSIONS
In this work, we have studied the nonequilibrium behavior
of autocatalytic networks by relying on the topological features of these systems. One of the key results of this study
is that, while the condition for autocatalysis is topological,
it allows for an analysis of the dynamics of autocatalytic
networks. Hence, building on recent advancements in the
stoichiometric analysis of autocatalysis, we have developed
a framework to describe their dynamics without requiring
steady-state conditions or specific reaction kinetics.
By exploiting the hypergraph representation of an autocatalytic network, we derived a general decomposition of
014414-11
ARMAND DESPONS
PHYSICAL REVIEW E 111, 014414 (2025)
chemical fluxes that is both broad and robust, revealing
fundamental insights into their operation far from equilibrium. Furthermore, the decomposition of chemical fluxes
into productive modes and cycles, based solely on the network’s stoichiometric matrix, highlighted two distinct types
of behavior: Productive fluxes that generate autocatalysts in
excess, and cyclic fluxes that preserve the internal state of
the network. This separation, dictated purely by topology,
emphasizes the powerful relationship between structure and
dynamics in these systems. In addition, the structure of these
networks, particularly through the examination of conservation laws, is sufficient to infer the physical roles played by
different chemical species.
A significant aspect of our analysis is the role of external species coupled with the autocatalytic networks. Indeed,
stoichiometric autocatalysis, by definition, violates mass conservation, meaning that the system requires external species,
which function as fuel or waste materials, enabling the autocatalytic network to operate. In the context of biology, these
species should be constantly provided to the autocatalytic
network and, in addition, external mechanisms must replenish
fuel species from the excess of waste. Even though maintaining these external species bring nonconservative forces, we
demonstrated that these forces do not apply on the productive modes of the autocatalytic networks. This was absent in
previous analysis of open chemical networks, in which the
nonconservative forces are considered homogeneously among
the reactions. As a result, the condition for unconditional
detailed balance in autocatalytic networks is independent of
the nonconservative forces brought by the external species.
The thermodynamic properties of autocatalytic networks
also exhibit distinct characteristics. From the nonequilibrium
geometry of autocatalytic networks, we derived a correction
that must be applied to the semigrand Gibbs free energy
to accurately describe the thermodynamics of autocatalytic
networks. This correction arises to take into account the underlying conservative process of external species sustaining
the production of autocatalysts. From the EPR decomposition
of the thermodynamic potential, and the decomposition of the
fluxes, we were able to distinguish the cost sustaining the
futile cycles of autocatalytic networks from the cost sustaining
the production of autocatalysts. While the former ends up
dissipated, the latter is able to generate useful work that can
be extracted or change the free-energy content in the autocatalytic network.
In summary, our results underscore the central role of
topology in governing both the dynamics and thermodynamics of autocatalytic networks. By linking conservation
laws, flux decomposition, and thermodynamic potentials, we
have provided a comprehensive framework for understanding
these systems in out-of-equilibrium conditions, with implications for both theoretical studies and practical applications
in biological and chemical contexts. Nevertheless, in the
metabolism, autocatalysis generally occurs inside a compartment. In that case, maintaining fluxes along the productive
modes will be induce a priori volume growth (or shrinkage).
In this work, we have neglected this effect, assuming constant volume. In this regard, Sughiyama et al. have recently
proposed an innovative framework addressing the chemical
thermodynamics of a growing system in Ref. [46]. However,
FIG. 5. The non-mass-like conservation laws in an autocatalytic
subnetwork can be traced back to the presence of creationannihilation motifs in the network.
it was restricted to the nonsingular stoichiometric matrix S;
we hope that the results derived here will help to extend
their results in the general case, leaving the extension of this
framework for future studies.
ACKNOWLEDGMENTS
The author acknowledges fruitful discussions with P. Raux,
A. Blokhuis, and D. Lacoste and further thanks Y. Sakref and
L. Dinis for their suggestions on an earlier draft.
APPENDIX A: GENERAL CASE
In the most general framework of Eq. (1), the left null
space of S is not empty, ker S = {0}; subsequently, the autocatalytic subnetwork has conservation laws. Still, Gordan’s
theorem [51] implies that the stoichiometric condition Eq. (1)
is equivalent to the absence of mass-like conservation laws in
the subnetwork. In other words, although not empty, the left
null space of S is not intersecting the positive orthant:
ker S ∩ R|Z|
>0 = ∅.
(A1)
The (non-mass-like) conserved quantities in autocatalytic networks are usually caused by creation-annihilation motifs.
Such a motif is represented in Fig. 5 and consists of a complex
of autocatalytic species that is formed and destroyed. For
example, for the motif represented in Fig. 5,
1 −1
0
= 0
Z2
···
· · · Z1
is associated with the conserved quantity L = [Z1 ] − [Z2 ],
which expresses that species Z1 and Z2 cannot be produced
independently. As we did with the conservation laws of the
full network, we can gather a linear basis of the left null space
of S as the rows of a matrix L:
L · S = 0.
(A2)
1. Autocatalytic potential species
Exchanging autocatalytic species with the environment is
likely to break some of the non-mass-like conservation laws
of the subnetwork. For example, if the motif in Fig. 5 is
present in the subnetwork, its conserved quantity is broken
once species Z1 (or Z2 ) is exchanged.
As a result, some of the species in Y will be “potential”
species, associated with the broken conservation laws of the
subnetwork. We denote by Y p ⊂ Y the subset of the autocatalytic potential species, and we let Y f = Y − Y p be the subset
014414-12
NONEQUILIBRIUM PROPERTIES OF AUTOCATALYTIC …
PHYSICAL REVIEW E 111, 014414 (2025)
of the remaining autocatalytic force species. The rows of L
associated with species Y p , L Y p , are the broken conservation
laws of the subnetwork, and, without loss of generality, we
directly work with its row-reduced form:
Y
Y
(A3)
L Y p = 1|Y p | , −LY pf , −LXp .
2. Broken conservation laws
associated with the unbroken conservation laws of the subnetwork. Setting a nonzero external flux on species x p ∈ X p
would result in a new broken conservation law.
In the end, one has Z p = Y p ∪ X p while the remaining
autocatalytic species are Z f ≡ Z − Z p . Importantly, the autocatalytic force species verify Y f ⊂ Z f , and finally, X f ≡
Z f − Y f ⊂ X . Consequently, the stoichiometric matrix of the
subnetwork has the following block decomposition,
The broken conservation laws of the network are now
obtained by considering L and L Y p :
(A6)
(A4)
such that S is deprived of conservation law: ker S = {0}. On
the other hand, the conservation laws of S are associated with
species Z p ,
L = 1|Z p | , −LZ f .
Its restriction to the potential species, p = E p ∪ Y p , is
⎞
⎛
LY p
LE p
⎟
⎜
⎟,
Lp = ⎜
⎠
⎝
0
1|Y p |
which is a non-singular matrix:
⎛ −1
LE p
⎜
L p −1 = ⎜
⎝
0
−MY p
Crucially, the condition in Eq. (A1) implies that each column
of LZ f has at least one strictly positive entry:
z
(A7)
∀z p ∈ Z p , ∃z f ∈ Z f such that LZ f z pf > 0.
⎞
As a result, the stoichiometry of species Z p , T , is directly
related to S:
⎟
⎟.
⎠
T = LZ f · S.
1|Y p |
Hence, the moiety matrix associated with all the potential
species reads
Finally, we obtain elementary modes of the species in Z f by
taking a right inverse of S,
S · G = 1|Z f | .
(A5)
(A8)
(A9)
In addition, using Eq. (A8), the change in species Z p along
the productive modes is
T · G = LZ f .
(A10)
Therefore, Eq. (A7) insures that, for all z p ∈ Z p and for every
choice of right inverse G, there exists a species z f ∈ Z f whose
elementary mode, gz f , is also producing species z p :
where
Y
MY f = MY f + MY p · LY pf ,
z
T · gz f p > 0.
Y
MX = MX + MY p · LXp .
3. Elementary modes of production
The presence of conservation laws in the subnetwork
prevents the definition of elementary modes for all the
autocatalytic species. Specifically, we can relate the conservation laws of the subnetwork to a subset Z p ⊂ Z of the
autocatalytic species. Because they are associated with the
broken conservation laws, the autocatalytic potential species
necessarily belong to this subset, Y p ⊂ Z p ; yet, in addition,
the subnetwork might still have unbroken conservation laws.
These are related to the autocatalytic species that are not
exchanged with the environment, namely the X species. As
a result, we can define the subset of species X p ⊂ X that are
(A11)
Hence, when the most general criterion in Eq. (1) applies but
not its stricter counterpart Eq. (2), we can produce only a
subset Z f of the autocatalytic species independently, while
the remaining species Z p , which bear the non-mass-like conservation laws, are subjected to the production of species
in Z f . When Z f = Z and Z p = ∅, the subnetwork has no
conservation law and the stricter condition used in the main
text holds.
Remark. At this point, the notations X p and X f are used
for convenience because the species in these subsets are not
exchanged with the environment, and, hence, are neither “potential” nor “force.” Nevertheless, as shown in Appendix B
(cf. the remark), at equilibrium the species in X p (resp. X f )
behave as if they were potential (resp. force) species.
014414-13
ARMAND DESPONS
PHYSICAL REVIEW E 111, 014414 (2025)
As a result, when
4. Decomposition of the fluxes
The decomposition of the stoichiometric matrix in Eq. (A6)
imposes that
ker S = ker S.
(A12)
Consequently, Sec. II C of the main text is unaffected by
the presence of conservation laws in the subnetwork and its
results apply straightforwardly. In particular, when G R2 = 0,
the columns of (G, C) define a linear basis of the chemical
fluxes, and the decomposition in Eq. (28) applies. Nevertheless, in that case, the fluxes along the productive modes are
defined only on the subset of species Z f (instead of Z):
J ε (t ) cε +
j=
ε
J z f (t ) gz f .
(A13)
z f ∈Z f
E
R
=0
∇
2
and
Y f = ∅,
G relaxes to its minimum. Furthermore, when Y f = ∅ one
has X f = Z f , and the optimum of G with respect to the
autocatalytic species that are not externally controlled, i.e.
X = X p ∪ X f = X p ∪ Z f , is
−1
eq E
·
T
· SR1 ,
μeq
=
−
μ
·
∇
+
μ
,
μ
E
Y
R
Xf
Xp
p
1
μeq
X p = μE p · MX p ,
(B4)
which can also be obtained by balancing the chemical potentials. As before, equilibrium is the global minimum of G:
When the subnetwork approaches steady state,
G = G(x, y p , e) − G(xeq , y p , e) = L(x xeq ) ⩾ 0.
∀x f ∈ X f , J x f (t ) −→ 0,
∀y f ∈ Y f , J y f (t ) −→ J y f = −I y f ,
Remark. Plugging the equilibrium state in the nonconservative forces yields
such that the decomposition of the steady fluxes becomes
j = C · J + GY f · J Y f .
(A14)
APPENDIX B: THERMODYNAMICS OF THE
SUBNETWORK IN THE GENERAL CASE
F Xeqp = 0,
In the general case, the moieties exchanged with environment are
(B1)
In addition, the fundamental nonconservative forces read
F = μ − μ p · M,
(B5)
Hence, at equilibrium, the species in X p (resp. X f ) behave as
if they were potential (resp. force) species, having vanishing
(resp. nonvanishing) nonconservative forces.
1. Thermodynamic potential
m(e, z) = ME · [e] + MZ · [z].
−1
F Xeqf = F E · ∇ ER2 · SR1 .
(B2)
where μ p ≡ (μE p , μY p ) stores the chemical potentials of the
potential species p = E p ∪ Y p . As before, the nonconservative
forces of the potential species vanish: F E p = 0 and F Y p = 0.
Finally, the thermodynamic potential is given by Eq. (56),
APPENDIX C: PROOF OF EQUATION (42)
Upon setting nonvanishing external fluxes on all the external species one has [25]
|E| = #Emergent cycles in∇ Z + #Broken conservation laws.
(C1)
By definition, the last term is |E p |; hence, we are left with the
number of emergent cycles of
G(z, e) = G(z, e) − μ p · m + F E · e(z f ).
Note that, now, the conservative influx of external species, e,
depends only on the species in Z f ,
(C2)
e(z f ) = ∇ ER · G · z f .
2. Autonomous subnetworks
When the driving work rate vanishes, Ẇdriv = 0, the EPR
decomposition is similar to Eq. (63); yet, in the presence
of conservation laws in the subnetwork, the work rate along
the productive modes is expressed only on the subset of the
autocatalytic force species:
Ẇprod = −
G gy f I y f .
(B3)
y f ∈Y f
These are the cycles of ∇ Z that are not also cycles of ∇.
Clearly, performing once any of the additional reactions defines an emergent cycle but, in addition, the cycles of S
that are not cycles of the full network also suit. There is
E | of such cycles; thus
|ker S| − |ker ∇
R2
ER + |\R|.
|E f | = |ker S| − ker ∇
2
Noticing that, |ker S| = |R2 | yields Eq. (40).
014414-14
(C3)
NONEQUILIBRIUM PROPERTIES OF AUTOCATALYTIC …
PHYSICAL REVIEW E 111, 014414 (2025)
APPENDIX D: DETAILS FOR FIG. 1
The stoichiometric matrix of the network in Fig. 1(a) is
(D1)
where the black line tells the subset of external species E (top) and the subset of autocatalytic species Z (bottom) apart. Because
there is no additional reaction in this network (\R = ∅), the upper block is ∇ ER , while the lower block is the autocatalytic
subnetwork S. Indeed, the latter fulfills the stoichiometric condition Eq. (1) because the linear combination of reactions given by
u in Fig. 1(b) creates a net excess of each species in the subset Z: S · u = 2 > 0 (component-wise). Additionally, the network
has six independent conservation laws (| ker ∇ | = 6):
(D2)
which are all mass-like and brought by the external species. As a result, the autocatalytic subnetwork is deprived of conservation
laws and, on top of the general criterion, S fulfills the stronger condition Eq. (2). According to the choice of potential species in
the main text, the moiety matrix reads
(D3)
The first row locates the phosphate groups exchanged among the species, while the second row locates the fragments made of
6 carbon atoms which are brought by glucose. Finally, as expected, on the subset of potential species, the moiety matrix is the
identity matrix: ME p = 1|E p | .
[1] P. Schuster, Monatsh. Chem. 150, 763 (2019).
[2] A. Roy, D. Goberman, and R. Pugatch, Proc. Natl. Acad. Sci.
USA 118, e2107829118 (2021).
[3] W.-H. Lin, E. Kussell, L.-S. Young, and C. Jacobs-Wagner,
Proc. Natl. Acad. Sci. USA 117, 27795 (2020).
[4] Y. Sakref and O. Rivoire, J. Theor. Biol. 579, 111714 (2024).
[5] S. Ameta, Y. J. Matsubara, N. Chakraborty, S. Krishna, and S.
Thutupalli, Life 11, 308 (2021).
[6] Z. Peng, J. Linderoth, and D. A. Baum, PLoS Comput. Biol. 18,
e1010498 (2022).
[7] W. Hordijk and M. Steel, Origins Life Evol. Biosphere 44, 111
(2014).
[8] L. Vincent II, S. Colón-Santos, H. J. Cleaves, D. A. Baum, and
S. E. Maurer, Life 11, 1221 (2021).
[9] Z. Peng, A. M. Plum, P. Gagrani, and D. A. Baum, J. Theor.
Biol. 507, 110451 (2020).
014414-15
ARMAND DESPONS
PHYSICAL REVIEW E 111, 014414 (2025)
[10] J. C. Xavier, W. Hordijk, S. Kauffman, M. Steel, and W. F.
Martin, Proc. R. Soc. B 287, 20192377 (2020).
[11] P. G. Higgs and N. Lehman, Nat. Rev. Genet. 16, 7 (2015).
[12] H. S. Bernhardt, Biology Direct 7, 23 (2012).
[13] P. Pavlinova, C. N. Lambert, C. Malaterre, and P. Nghe, FEBS
Lett. 597, 344 (2023).
[14] S. A. Kauffman, J. Cybernetics 1, 71 (1971).
[15] S. A. Kauffman, J. Theor. Biol. 119, 1 (1986).
[16] A. Blokhuis, D. Lacoste, and P. Nghe, Proc. Natl. Acad. Sci.
USA 117, 25230 (2020).
[17] J. L. Andersen, C. Flamm, D. Merkle, and P. F. Stadler,
arXiv:2107.03086.
[18] J. W. Gibbs, The Collected Works of J. Willard Gibbs, Vol. 1,
Thermodynamics (Longmans, Green, and Co., London, 1928).
[19] T. de Donder, L’affinité (Gauthier-Villars, Paris, 1927).
[20] D. A. McQuarrie, J. Appl. Probab. 4, 413 (1967).
[21] D. T. Gillespie, Phys. A 188, 404 (1992).
[22] J. Schnakenberg, Rev. Mod. Phys. 48, 571 (1976).
[23] C. Y. Mou, J.-L. Luo, and G. Nicolis, J. Chem. Phys. 84, 7011
(1986).
[24] T. L. Hill, Proc. Natl. Acad. Sci. USA 80, 2922 (1983).
[25] R. Rao and M. Esposito, Phys. Rev. X 6, 041064 (2016).
[26] H. Ge and H. Qian, Chem. Phys. 472, 241 (2016).
[27] F. Avanzini, E. Penocchio, G. Falasco, and M. Esposito,
J. Chem. Phys. 154, 094114 (2021).
[28] R. A. Alberty, Thermodynamics of Biochemical Reactions
(Wiley, New York, 2003).
[29] H. Qian and D. A. Beard, Biophys. Chem. 114, 213 (2005).
[30] S. Dal Cengio, V. Lecomte, and M. Polettini, Phys. Rev. X 13,
021040 (2023).
[31] P. E. Harunari, A. Dutta, M. Polettini, and Édgar Roldán, Phys.
Rev. X 12, 041026 (2022).
[32] A. Despons, Y. de Decker, and D. Lacoste, Commun. Phys. 7,
224 (2024).
[33] T. Aslyamov and M. Esposito, Phys. Rev. Lett. 132, 037101
(2024).
[34] J. A. Owen and J. M. Horowitz, Nat. Commun. 14, 1280
(2023).
[35] M. Feinberg, Foundations of Chemical Reaction Network Theory (Springer International Publishing, Berlin, 2019).
[36] I. R. Epstein and J. A. Pojman, An Introduction to Nonlinear Chemical Dynamics (Oxford University Press, New York,
1998).
[37] T. G. Kurtz, J. Chem. Phys. 57, 2976 (1972).
[38] H. Ge and H. Qian, Phys. Rev. E 94, 052150 (2016).
[39] M. Pekař, Prog. React. Kinet. Mech. 30, 3 (2005).
[40] R. Rao and M. Esposito, J. Chem. Phys. 149, 245101 (2018).
[41] H. S. Haraldsdóttir and R. M. T. Fleming, PLoS Comput. Biol.
12, e1004999 (2016).
[42] F. Avanzini and M. Esposito, J. Chem. Phys. 156, 014116
(2022).
[43] D. L. Nelson and M. Cox, Principles of Biochemistry (W. H.
Freeman and Co., New York, 2024), Chap. 14.
[44] M. Polettini and M. Esposito, J. Chem. Phys. 141, 024117
(2014).
[45] A. Wachtel, R. Rao, and M. Esposito, New J. Phys. 20, 042002
(2018).
[46] Y. Sughiyama, A. Kamimura, D. Loutchko, and T. J. Kobayashi,
Phys. Rev. Res. 4, 033191 (2022).
[47] F. Avanzini, N. Freitas, and M. Esposito, Phys. Rev. X 13,
021041 (2023).
[48] P. Raux, C. Goupil, and G. Verley, Phys. Rev. E 110, 014134
(2024).
[49] A. Wachtel, R. Rao, and M. Esposito, J. Chem. Phys. 157,
024109 (2022).
[50] T. M. Cover and J. A. Thomas, Elements of Information Theory
(Wiley, New York, 2005).
[51] P. Gordan, Math. Ann. 6, 23 (1873).
014414-16