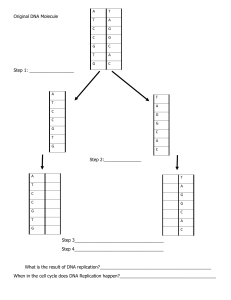
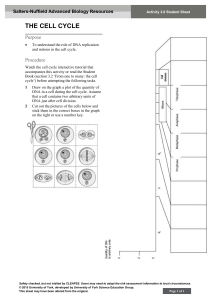
Biol 2100 learning objectives The best way to engage with these learning objectives is to attempt them (explain, predict, describe, draw, calculate, interpret, etc.) with a classmate. Only someone who is familiar with the content may recognize when you’ve been incomplete or incorrect. You should be able to do these things without referring to your notes, textbook, or lecture recordings. Identifying early what you’re not yet capable of doing provides an opportunity to get help and achieve that objective before the exam! Exam 1 Chemical foundations essential to understanding cellular and molecular biology • Explain why the distinction between covalent and non-covalent is important to biologists, and what distinguishes covalent bonds from ionic bonds, hydrogen bonds, van der Waals and hydrophobic interactions • Explain the relationship between polar covalent bonds and hydrogen bonds • Predict hydrophilicity/hydrophobicity based on chemical structure • Recognizing an association as being covalent or non-covalent based on an image or description Protein structure and function • Draw the general structure of an amino acid in both ionized and non-ionized forms • Recognize by structure amino acids with highly hydrophobic or highly hydrophilic side chains • Draw or model the effect of pH on amino acids and relation to protein structure and function • Identify hydroxyl, carbonyl, carboxyl, amino, sulfhydryl, phosphate, and methyl groups • Define monomer, residue, polymer, peptide, polypeptide chain, and protein • Define hydrolysis and dehydration/condensation reactions and model with a dipeptide • Describe, identify, and note stabilizing forces of primary, secondly, tertiary, & quaternary structure • Describe typical location of hydrophobic and hydrophilic sidechains in a protein • Illustrate the principles of structural and functional domains (new) • Calculate the number of molecules in a particular volume based on concentration in mass/volume • Interpret SDS-PAGE data Cell structure and cytoskeleton • Distinguish cytoplasm and cytosol • Interpret immunofluorescent microscopy images • Describe the structure and functions of microtubules, microfilaments, and intermediate filaments • Recognize names of proteins forming microtubules, microfilaments, and associated motor proteins • Explain and illustrate the concept of structural polarity and its significance to cytoskeletal filaments • List similarities and differences between plant, animal, and prokaryotic cells Lipids and membranes • Recognize membrane lipids by their structures, explaining how and why they are amphipathic • Give an example of the significance of the asymmetric distribution of membrane phospholipids • Relate lipid structural variations to membrane permeability and describe features of solutes that influence their ability to diffuse across cell membranes • Recognize cis- versus trans- double bonds, explaining which is in our membrane lipids and why 1 Membrane transport, endocytosis and exocytosis • Recognize integral (transmembrane), peripheral, and lipid-linked membrane proteins • Distinguish channels, uniporters, symporters, antiporters, and pumps; describe mechanisms driving active transport (both primary and secondary/indirect) • Predict the direction of osmosis, appropriately using the relative terms hypertonic and hypotonic • Explain the effect of electrochemical gradients on the diffusion of solutes • Distinguish phagocytosis, pinocytosis, and receptor-mediated endocytosis from one another and from membrane transport of solutes Free energy and coupled reactions • Define and relate “free energy”, exergonic, endergonic, entropy, enthalpy, endo- and exothermic • Describe the influence of enthalpy and entropy changes on DG of a reaction or process • Explain the principle of coupled reactions using a biological example • Given DG values, determine whether reactions may be effectively coupled • Describe what DG of a reaction or process does, and does not, tell us Enzymes • Explain why thermodynamically favorable reactions won’t occur instantaneously • Define enzyme, explain why cells use enzymes rather than temperature to regulate reaction rates • Explain why enzymes do not change the DG of a reaction • Describe examples of how enzymes function including the role of co-factors • Explain Vmax, Km and how each is influenced by competitive and non-competitive inhibitors • Explain why different enzymes have different optimal temperature and pH activity profiles 2 Exam 2 Cellular respiration and fermentation • Define oxidation and reduction; identify oxidizing and reducing agents in redox reactions • Remember the oxidized/reduced states of NAD+/NADH (and which has greater free energy) • Describe the flow of energy and recycling of chemicals between photosynthesis and cellular respiration, noting the overall change in oxidation state of carbon in these processes • Explain why and how ATP connects energy harvesting and energy requiring processes • Describe the purposes of cellular respiration • Remember the location and inputs/outputs of glycolysis, pyruvate oxidation, Krebs cycle, and the relative net ATP production of each phase per input molecule • Distinguish ATP production by substrate level phosphorylation versus by oxidative phosphorylation • Explain the role of molecular oxygen in aerobic respiration and how anaerobic respiration differs • Remember the relative ATP value of NADH and FADH2 in oxidative phosphorylation • Describe the location, functioning and purpose of the mitochondrial electron transport chain • Explain the common purpose of fermentation pathways, clearly distinguishing fermentation from anaerobic cellular respiration • Explain how ATP can act as both a substrate and as an inhibitor of phosphofructokinase Photosynthesis • Explain the relationship between photosynthesis and cellular respiration • Describing the purpose of, and relationship between, the light reactions and the Calvin cycle • Note the key inputs, outputs, and precise location of the light reactions and the Calvin cycle Cell signaling • Define and recognize endocrine, paracrine, autocrine, juxtacrine, and synaptic signaling • Describe key features of steroid, ligand-gated ion channel, receptor tyrosine kinase, and G-protein coupled receptor signaling, including their activation and termination • Describe the activity of kinases and phosphatases and effects of phosphorylation on protein targets • Describe the principle of signal transduction and how it occurs in receptor tyrosine kinase and Gprotein coupled receptor signaling • Describe the function of G-proteins and mechanisms of their activation and inactivation • Describe and recognize signal amplification • Define second messenger, note their defining characteristics, and explain how they act • Remember the activities of adenylyl cyclase and phosphodiesterase • Recognize components of cell signaling pathways (receptors, ligands, adapters, second messengers) • Define apoptosis and given an example of when it occurs • Interpret cellular pathway diagrams involving symbols for activation/stimulation and inhibition/blockade Mitosis and the cell cycle • Describe connections between cell signaling and cell division • Describe how the daughter cells of mitosis compare to the parent cell • Provide examples of when, where, and why mitosis occurs • Describe defining features of each phase (G1/G0 S, G2, M) of the cell cycle • Explain how the sequence and duration of cell cycle phases can be determined experimentally • Note the defining features of prophase, metaphase, anaphase, telophase and cytokinesis, and explain the significance of those defining features 3 Cell cycle regulation • Describe the activity of cyclin/Cdk complexes in driving cell cycle transitions; with the example of MPF, relate its activity to the defining features of mitotic phases • Describe examples of cell cycle “checkpoints”, distinguishing from Cdk/cyclin complexes • Describe the importance and key functions of p53 • Describe the consequences of different cell cycle checkpoint failures in mitosis 4 Exam 3 Meiosis • Explain the purpose and location of meiosis, fundamentally distinguishing meiosis from mitosis • Illustrate or model the process of meiosis with at least two different chromosome pairs and two genes per chromosome (incorporate crossing-over and precisely distinguish meiosis I and II) • Distinguish recombinant and non-recombinant chromosomes • Define haploid, diploid, polyploid, aneuploid. Distinguish a trisomy from triploidy • Contrast the processes and outcomes of meiosis and mitosis • Describe how failures in meiosis can result in chromosomal abnormalities Mendelian inheritance • Describe key features of Mendel’s experimental model and the characters that he studied • Explain the evidence against a blending hypothesis of inheritance • Define dominant and recessive, phenotype and genotype • Illustrate Mendel’s evidence for independent rather than dependent assortment • Relate Mendel’s principles of segregation and independent assortment to the process of meiosis • Correctly apply the rules of multiplication and addition in calculating probabilities Extending beyond Mendel • Distinguish and recognize codominance versus incomplete dominance • Explain and distinguish multiple allelism and polygenic inheritance • Describe incomplete penetrance and explain why we might observe it • Describe epistasis, pleiotropy, and quantitative effects Connecting genes to chromosomes • Properly interpret a pedigree chart • Recognize autosomal, X-linked, Y-linked and mitochondrial gene inheritance patterns • Explain how expression of heterozygosity is fundamentally different between autosomal and Xchromosome genes in female mammals Linked genes • Describe why particular alleles of two different genes would not assort independently • Explain how and why recombination frequency between genes reflects the relative distance between those genes on the chromosome (relating back to the process of meiosis) • Explain how a mating experiment can be designed to reflect recombinants from just one parent • Properly identify recombinant versus non-recombinant offspring relative to gene pairs and map the relative position of genes based on recombination frequency Linking genes to the DNA of chromosomes • Explain Griffith’s remarkable observation of bacterial transformation • Describe how Avery demonstrated that Griffith’s transforming factor is DNA • Explain how Hershey and Chase could track DNA and protein separately, and their major findings 5 Exam 4 Nucleotides • Describe the general structure of nucleosides and nucleotides, distinguishing those of DNA from those of RNA, and remembering which are purines and which are pyrimidines • Remember what NTP and dNTP stand for, and which bases are included in each • Remember the significance of the 1’, 2’, 3’ and 5’ positions of ribose and deoxyribose • Identify the 3’ → 5’ or 5’ → 3’ directionality of a nucleotide or nucleic acid strand • Explain why nucleic acid polymerization (synthesis) is 5’ → 3’ • Summarize the major discoveries of Chargaff, Franklin, and Watson & Crick DNA replication • Illustrate the principle of semi-conservative replication of DNA with a simple drawing • Illustrate Meselson and Stahl’s predictions based on three hypotheses of DNA replication including their actual results and evidence for semi-conservative replication (dump this?) • Explain why an RNA polymerase is required for DNA replication • Draw an origin of replication, replication forks, and illustrate bidirectional continuous and discontinuous DNA synthesis in the correct direction relative to the template strands, including the activities of Primase, DNA polymerases (precisely noting what serves as primers), and DNA ligase • Summarize the functions of helicase, single-strand binding proteins and topoisomerase • Explain when, where and why telomerase activity is required and draw its activity (clearly illustrating that it is an RNA-dependent DNA polymerase) Transcription and RNA processing • Contrast DNA replication with transcription (purpose, location, enzymes, and nucleotides) • Name and describe the DNA sequences that begin and end transcription • Describe the relationships between DNA template, non-template, and coding strand • Describe the major features and purposes of pre-mRNA processing in eukaryotes • Define intron and exon and relate alternative splicing to protein structure and function Translation • Explain the role of tRNA molecules and aminoacyl tRNA synthetases • Describe where codons and anticodons are found and their role • Describe one function of an mRNA UTR (untranslated region) (dump this?) • Remember ATG/AUG as the start and methionine codon, and that stop codons are recognized by a protein release factor (not a tRNA) • Explain the concept of reading frame and properly use a genetic code chart/table • Explain how the genetic code is redundant, yet not ambiguous • Explain and distinguish the two mechanisms resulting in genetic code redundancy • Describe and distinguish silent (synonymous), missense, nonsense, and frameshift mutations Protein synthesis and targeting in eukaryotic cells • Explain what is meant by the term “secretory protein”, what feature distinguishes “secretory” from “non-secretory” proteins, and how that difference regulates protein trafficking in eukaryotic cells • Explain the location, role, and effect of signal peptidase • Remember that folding, N-linked glycosylation, & quaternary structure are initiated in the ER lumen • Explain how the presence/absence of a transmembrane anchor influences protein targeting 6 • For a transmembrane protein, illustrate the locations of the intracellular and extracellular domains as that protein moves from ER to Golgi to plasma membrane • Explain what determines whether a non-secretory protein will remain in the cytosol, traffic to the nucleus, or be imported into another intracellular compartment PCR (move entirely to lab, including Virtual Bacterial ID lab, not using lecture time) • Explain what’s occurring in each step of a PCR cycle, noting the purpose and fate of the primers and why some DNA products are longer than intended (or desired) • Given a target sequence, recognize a primer pair that could be used for amplification • Predict the length of PCR products given primer and target sequence information • Compare and contrast DNA replication in cells with DNA synthesis in PCR Dideoxy DNA sequencing (as above with PCR) • Explain what a dideoxy nucleotide is and effect of its presence during DNA synthesis • Note the required components of an automated (fluorescent) DNA sequencing reaction • Properly interpret a sequencing chromatogram (does it illustrate the sequence of the template or of the synthesis product? From 5’ to 3’ or 3’ to 5’? What differs between adjacent peaks? What does the area under the curve represent? What do overlapping peaks mean?) Exam 5 Regulation of gene expression in prokaryotes • Describe how replica plating can be used to identify certain lac operon mutants • Explain what a “constitutive” mutant is (introducing the idea of negative control) • Explain how “partial diploid” mutants provide evidence for negative control acting through a combination of a protein (repressor) and a DNA sequence (operator) • Describe the components and characteristics of operons • Explain how and why glucose levels influence the lac operon (introducing positive control) • Distinguish and recognize positive versus negative control and explain the value of the combination • Relate environmental conditions to the state of the lac operon • Explain the fundamental similarities and differences between the lac and trp operons Regulation of gene expression in eukaryotes • Explain how DNAase-susceptibility experiments provide evidence of chromatin structure at particular loci, interpret such experimental data, and predict chromatin structure in relation to gene expression (dump this, and its required explanation of the Southern blot?) • Describe the typical effect of methylation of promotor CpG islands • Explain what genomic imprinting is and its effect on somatic cell gene expression • Define distal and proximal; explain the role of enhancers and promotor proximal elements in tissuespecific and coordinated gene expression • Explain how the iron response element example illustrates UTR regulation of gene expression at the levels of translation and mRNA stability (dump this?) • Describe what RNA-interference is, ways in which microRNAs (miRNA) can silence gene expression, and a natural role of miRNA Modifying organisms—transgenics and gene knockouts 7 • Describe what a (conventional) transgenic organism is, reasons why they are made, and why breeding is required to produce a “true-breeding” colony of transgenic animals (reiterating principles of genetics), and how transgenic organisms fundamentally differ from gene therapy recipients • Describe what a gene “knockout” organism is, common reasons for producing them, the role of embryonic stem cells, powerful selection strategies in their production (dump this part?), what chimeric animals are, and the role of breeding in producing homozygous gene KOs • Describe what “conditional” gene knockouts are, their significance, the role of the bacteriophage Cre-Lox system in their generation, what “floxed”, “knock-in”, and hemizygous mean, and relate their production to basic Mendelian genetics CRISPR/Cas and gene therapy (perhaps least essential, but interesting and course-wide integrative) • Describe what CRISPR and Cas9 are, their natural function and location, and how CRISPR targets Cas9 • Explain which parts of CRISPR/Cas are typically being exploited to alter eukaryotic genes and the required modification to Cas9 • Distinguish non-homologous end joining (NHEJ) and homology directed repair (HDR), noting their significance following Cas9 activity • Understand examples of Cas9 gene editing, interpreting studies involving methods and principles that you’ve learned in Bio 2100 • Explain the challenges that remain in exploiting Cas9 editing in humans Introduction to stem cells and regenerative medicine (dump?) • describe what iPSCs (induced pluripotent stem cells) are and how their therapeutic potential complements that of gene therapy • analyze/interpret Western blot data (this may be integrated elsewhere) class and lab activities will target the most challenging of these (exams 1,2,4 had raw ave of ~70%, 3 and 5 were ~78%) 8