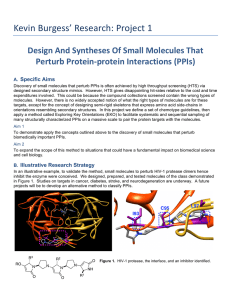
Protein-Protein Interactions Within Cells: Mechanisms and Implications Abstract Protein-protein interactions (PPIs) are fundamental to cellular processes, influencing everything from signal transduction to structural organization. This paper reviews the mechanisms underpinning PPIs, the methodologies for studying these interactions, and their implications in both health and disease. Understanding PPIs is crucial for advancing biomedical research, with potential applications in drug discovery and therapeutic interventions. Introduction Proteins are essential macromolecules that execute a myriad of functions within biological systems. Their interactions with each other are critical for maintaining cellular homeostasis and regulating biochemical pathways. PPIs can be transient or stable, specific or promiscuous, and are central to cellular architecture and function. The study of PPIs provides insights into the molecular basis of diseases and opens avenues for targeted drug development. Mechanisms of Protein-Protein Interactions Structural Basis of PPIs PPIs occur through various types of interactions including hydrogen bonds, ionic bonds, hydrophobic interactions, and van der Waals forces. These interactions are often facilitated by specific domains or motifs within the protein structures: SH2 and SH3 domains: Recognize phosphotyrosine-containing and proline-rich sequences, respectively, and are crucial in signal transduction pathways. PDZ domains: Bind to C-terminal sequences of target proteins, playing roles in scaffolding and signaling complexes. Leucine zippers and helix-loop-helix motifs: Mediate dimerization and are important in transcription factor function. Types of PPIs PPIs can be classified based on their duration and function: Transient interactions: These are often involved in signaling pathways where proteins interact briefly to transmit signals. Examples include kinase-substrate interactions. Stable interactions: These form lasting complexes such as those in ribosomes or proteasomes, which are essential for protein synthesis and degradation, respectively. Allosteric Modulation PPIs can induce conformational changes that affect protein activity. Allosteric modulation is a mechanism where binding at one site on a protein affects the function at a different site. This can either enhance or inhibit the protein’s activity, playing a critical role in metabolic regulation and signal transduction. Methods for Studying Protein-Protein Interactions Experimental Techniques Co-immunoprecipitation (Co-IP): This technique uses antibodies to precipitate a protein of interest along with its interacting partners from cell lysates, allowing the study of complex formation. Yeast Two-Hybrid (Y2H) System: This genetic approach identifies interactions by detecting the activation of reporter genes in yeast cells. Fluorescence Resonance Energy Transfer (FRET): This technique measures energy transfer between two fluorescently labeled proteins to study their interactions in live cells. X-ray Crystallography and Nuclear Magnetic Resonance (NMR) Spectroscopy: These methods provide high-resolution structural details of protein complexes. Computational Approaches Molecular Docking: Predicts the preferred orientation of one protein when bound to another, helping to infer interaction sites. Molecular Dynamics Simulations: Provides insights into the dynamic nature of PPIs and conformational changes over time. Bioinformatics Tools: Databases like STRING and IntAct compile known PPIs, while algorithms predict potential interactions based on protein sequences and structures. Biological Implications of Protein-Protein Interactions Signal Transduction PPIs are pivotal in signaling cascades such as the MAPK pathway, where sequential interactions between kinases lead to cellular responses to external stimuli. Dysregulation of these interactions can result in diseases like cancer. Structural Integrity Proteins such as actin and tubulin interact to form cytoskeletal structures that maintain cell shape and facilitate intracellular transport. Mutations affecting these interactions can lead to structural disorders. Metabolic Regulation Enzyme complexes, such as those involved in glycolysis, are regulated through PPIs. Allosteric interactions within these complexes ensure efficient metabolic flux and adaptation to cellular energy demands. Disease Mechanisms Altered PPIs are implicated in numerous diseases. For instance, in neurodegenerative diseases like Alzheimer’s, aberrant interactions of tau protein and amyloid-beta lead to toxic aggregates. Understanding these interactions is crucial for developing therapeutic interventions. Therapeutic Applications Drug Discovery Targeting PPIs is a promising strategy in drug development. Small molecules or peptides designed to disrupt or stabilize specific interactions can modulate protein functions. For example, inhibitors of the Bcl-2/Bax interaction promote apoptosis in cancer cells, providing a therapeutic avenue for cancer treatment. Biomarker Identification PPIs can serve as biomarkers for disease states. The detection of specific protein complexes can aid in early diagnosis and monitoring of diseases. For instance, the interaction between prostate-specific antigen (PSA) and its binding proteins is used in prostate cancer diagnostics. Conclusion Protein-protein interactions are integral to cellular function, influencing diverse biological processes and disease mechanisms. Advances in experimental and computational techniques have significantly enhanced our understanding of PPIs, providing insights into their roles in health and disease. The study of PPIs holds immense potential for therapeutic interventions, making it a critical area of biomedical research. Future efforts should focus on elucidating the dynamic nature of these interactions and developing innovative strategies to modulate them for therapeutic benefit. References Pawson, T., & Scott, J. D. (1997). Signaling through scaffold, anchoring, and adaptor proteins. Science, 278(5346), 2075-2080. Nooren, I. M., & Thornton, J. M. (2003). Diversity of protein-protein interactions. The EMBO Journal, 22(14), 3486-3492. Berg, J. M., Tymoczko, J. L., & Stryer, L. (2002). Biochemistry. W H Freeman. Perkins, J. R., et al. (2010). Transient protein-protein interactions: structural, functional, and network properties. Structure, 18(10), 1233-1243. Keskin, O., Tuncbag, N., & Gursoy, A. (2016). Predicting protein–protein interactions from the molecular to the proteome level. Chemical Reviews, 116(8), 4884-4909.